A major pleiotropic QTL identified for yield components and nitrogen content in rice (Oryza sativa L.) under differential nitrogen field conditions
- PMID: 33079957
- PMCID: PMC7575116
- DOI: 10.1371/journal.pone.0240854
A major pleiotropic QTL identified for yield components and nitrogen content in rice (Oryza sativa L.) under differential nitrogen field conditions
Abstract
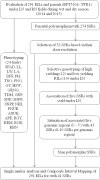
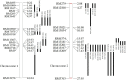
To identify the genomic regions for yield and NUE of rice genotypes and lines with promising yield under low N, a recombinant inbred population (RIL) developed between BPT5204 (a mega variety known for its quality) and PTB1 (variety with high NUE) was evaluated for consecutive wet and dry seasons under low nitrogen (LN) and recommended nitrogen (RN) field conditions. A set of 291 RILs were characterized for 24 traits related to leaf, agro-morphological, yield, N content and nitrogen use efficiency indices. More than 50 RILs were found promising with grain yield >10 g under LN. Parental polymorphism survey with 297 SSRs and selective genotyping revealed five genomic regions associated with yield under LN, which were further saturated with polymorphic SSRs. Thirteen promising SSRs were identified out of 144 marker trait associations under LN using single marker analysis. Composite interval mapping showed 37 QTL under LN with five pleiotropic QTL. A major stable pleiotropic (RM13201-RM13209) from PTB1 spanning 825.4 kb region associated with straw N % (SNP) in both treatments across seasons and yield and yield related traits in WS appears to be promising for the MAS. Another major QTL (RM13181-RM13201) was found to be associated with only relative trait parameters of biomass, grain and grain nitrogen. These two major pleiotropic QTL (RM13201-RM13209 and RM13181-RM13201) on chromosome 2 were characterized for their positive allele effect and could be deployed for the development of rice varieties with NUE.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Glass AD. Nitrogen use efficiency of crop plants: physiological constraints upon nitrogen absorption. Crit Rev Plant Sci. 2003. September 1;22(5): 453–470.
-
- Raun WR, Johnson GV. Improving nitrogen use efficiency for cereal production. Agron J. 1999; 91(3): 357–63.
-
- Vijayalakshmi P, Kiran TV, Rao YV, Srikanth B, Rao IS, Sailaja B, et al. Physiological approaches for increasing nitrogen use efficiency in rice. Indian J Plant Physiol. 2013. September 1; 18(3):208–22.
-
- Xia L, Zhiwei S, Lei J, Lei H, Chenggang R, Man W, et al. High/low nitrogen adapted hybrid of rice cultivars and their physiological responses. Afr J Biotechnol. 2011; 10(19): 3731–3738.
-
- Namai S, Toriyama K, Fukuta Y. Genetic variations in dry matter production and physiological nitrogen use efficiency in rice (Oryza sativa L.) varieties. Breed Sci. 2009; 59(3): 269–76.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

