Designed variants of ACE2-Fc that decouple anti-SARS-CoV-2 activities from unwanted cardiovascular effects
- PMID: 33080267
- PMCID: PMC7568492
- DOI: 10.1016/j.ijbiomac.2020.10.120
Designed variants of ACE2-Fc that decouple anti-SARS-CoV-2 activities from unwanted cardiovascular effects
Abstract
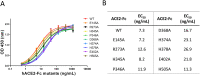
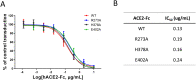
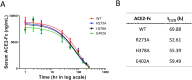
Angiotensin-converting enzyme 2 (ACE2) is the entry receptor for SARS-CoV-2, and recombinant ACE2 decoys are being evaluated as new antiviral therapies. We designed and tested an antibody-like ACE2-Fc fusion protein, which has the benefit of long pharmacological half-life and the potential to facilitate immune clearance of the virus. Out of a concern that the intrinsic catalytic activity of ACE2 may unintentionally alter the balance of its hormonal substrates and cause adverse cardiovascular effects in treatment, we performed a mutagenesis screening for inactivating the enzyme. Three mutants, R273A, H378A and E402A, completely lost their enzymatic activity for either surrogate or physiological substrates. All of them remained capable of binding SARS-CoV-2 and could suppress the transduction of a pseudotyped virus in cell culture. This study established new ACE2-Fc candidates as antiviral treatment for SARS-CoV-2 without potentially harmful side effects from ACE2's catalytic actions toward its vasoactive substrates.
Keywords: ACE2-Fc; COVID-19; Mutagenesis; SARS-CoV-2.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest Jing Jin and Pan Liu have applied for a provisional patent of the ACE2-Fc mutants described in the article.
Figures
Similar articles
-
Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig.Nat Commun. 2020 Apr 24;11(1):2070. doi: 10.1038/s41467-020-16048-4. Nat Commun. 2020. PMID: 32332765 Free PMC article.
-
Antiviral activity of an ACE2-Fc fusion protein against SARS-CoV-2 and its variants.PLoS One. 2025 Jan 3;20(1):e0312402. doi: 10.1371/journal.pone.0312402. eCollection 2025. PLoS One. 2025. PMID: 39752453 Free PMC article.
-
ACE2-based decoy receptors for SARS coronavirus 2.Proteins. 2021 Sep;89(9):1065-1078. doi: 10.1002/prot.26140. Epub 2021 May 18. Proteins. 2021. PMID: 33973262 Free PMC article. Review.
-
Picomolar inhibition of SARS-CoV-2 variants of concern by an engineered ACE2-IgG4-Fc fusion protein.Antiviral Res. 2021 Dec;196:105197. doi: 10.1016/j.antiviral.2021.105197. Epub 2021 Nov 10. Antiviral Res. 2021. PMID: 34774603 Free PMC article.
-
David versus goliath: ACE2-Fc receptor traps as potential SARS-CoV-2 inhibitors.MAbs. 2022 Jan-Dec;14(1):2057832. doi: 10.1080/19420862.2022.2057832. MAbs. 2022. PMID: 35380919 Free PMC article. Review.
Cited by
-
Utilizing noncatalytic ACE2 protein mutant as a competitive inhibitor to treat SARS-CoV-2 infection.Front Immunol. 2024 Apr 5;15:1365803. doi: 10.3389/fimmu.2024.1365803. eCollection 2024. Front Immunol. 2024. PMID: 38646520 Free PMC article.
-
Multivalent ACE2 engineering-A promising pathway for advanced coronavirus nanomedicine development.Nano Today. 2022 Oct;46:101580. doi: 10.1016/j.nantod.2022.101580. Epub 2022 Aug 4. Nano Today. 2022. PMID: 35942040 Free PMC article. Review.
-
His345 mutant of angiotensin-converting enzyme 2 (ACE2) remains enzymatically active against angiotensin II.Proc Natl Acad Sci U S A. 2021 Apr 13;118(15):e2023648118. doi: 10.1073/pnas.2023648118. Proc Natl Acad Sci U S A. 2021. PMID: 33833058 Free PMC article. No abstract available.
-
Functional Expression of the Recombinant Spike Receptor Binding Domain of SARS-CoV-2 Omicron in the Periplasm of Escherichia coli.Bioengineering (Basel). 2022 Nov 10;9(11):670. doi: 10.3390/bioengineering9110670. Bioengineering (Basel). 2022. PMID: 36354581 Free PMC article.
-
An ACE2 Triple Decoy that neutralizes SARS-CoV-2 shows enhanced affinity for virus variants.Sci Rep. 2021 Jun 17;11(1):12740. doi: 10.1038/s41598-021-91809-9. Sci Rep. 2021. PMID: 34140558 Free PMC article.
References
-
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. - PMC - PubMed
-
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. (e8) - PMC - PubMed
-
- Vickers C., Hales P., Kaushik V., Dick L., Gavin J., Tang J., Godbout K., Parsons T., Baronas E., Hsieh F., Acton S., Patane M., Nichols A., Tummino P. 277(17) 2002. Hydrolysis of Biological Peptides by Human Angiotensin-converting Enzyme-related Carboxypeptidase; pp. 14838–14843. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

