Diclofenac impairs autophagic flux via oxidative stress and lysosomal dysfunction: Implications for hepatotoxicity
- PMID: 33080439
- PMCID: PMC7575798
- DOI: 10.1016/j.redox.2020.101751
Diclofenac impairs autophagic flux via oxidative stress and lysosomal dysfunction: Implications for hepatotoxicity
Abstract
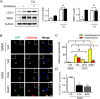
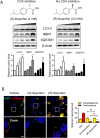
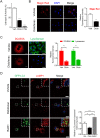
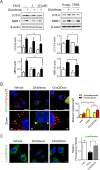
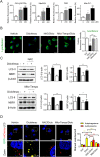
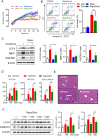
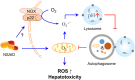
Treatment with nonsteroidal anti-inflammatory drugs (NSAIDs) is associated with various side effects, including cardiovascular and hepatic disorders. Studies suggest that mitochondrial damage and oxidative stress are important mediators of toxicity, yet the underlying mechanisms are poorly understood. In this study, we identified that some NSAIDs, including diclofenac, inhibit autophagic flux in hepatocytes. Further detailed studies demonstrated that diclofenac induced a reactive oxygen species (ROS)-dependent increase in lysosomal pH, attenuated cathepsin activity and blocked autophagosome-lysosome fusion. The reactivation of lysosomal function by treatment with clioquinol or transfection with the transcription factor EB restored lysosomal pH and thus autophagic flux. The production of mitochondrial ROS is critical for this process since scavenging ROS reversed lysosomal dysfunction and activated autophagic flux. The compromised lysosomal activity induced by diclofenac also inhibited the fusion with and degradation of mitochondria by mitophagy. Diclofenac-induced cell death and hepatotoxicity were effectively protected by rapamycin. Thus, we demonstrated that diclofenac induces the intracellular ROS production and lysosomal dysfunction that lead to the suppression of autophagy. Impaired autophagy fails to maintain mitochondrial integrity and aggravates the cellular ROS burden, which leads to diclofenac-induced hepatotoxicity.
Keywords: Diclofenac; Hepatotoxicity; Lysosomal dysfunction; Mitophagy; Nonsteroidal anti-inflammatory drugs; Reactive oxygen species.
Copyright © 2020 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures



References
-
- Gómez-Lechón M.J., Ponsoda X., O'Connor E. Diclofenac induces apoptosis in hepatocytes by alteration of mitochondrial function and generation of ROS. Biochem. Pharmacol. 2003;66(11):2155–2167. - PubMed
-
- Al-Attrache H., Sharanek A., Burban A. Differential sensitivity of metabolically competent and non-competent HepaRG cells to apoptosis induced by diclofenac combined or not with TNF-α. Toxicol. Lett. 2016;258:71–86. - PubMed
-
- Syed M., Skonberg C., Hansen S.H. Mitochondrial toxicity of diclofenac and its metabolites via inhibition of oxidative phosphorylation (ATP synthesis) in rat liver mitochondria: possible role in drug induced liver injury (DILI) Toxicol. Vitro. 2016;31:93–102. - PubMed
-
- Li H., Hortmann M., Daiber A. Cyclooxygenase 2-selective and nonselective nonsteroidal anti-inflammatory drugs induce oxidative stress by up-regulating vascular NADPH oxidases. J. Pharmacol. Exp. Therapeut. 2008;326(3):745–753. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

