Stat2-Drp1 mediated mitochondrial mass increase is necessary for pro-inflammatory differentiation of macrophages
- PMID: 33080440
- PMCID: PMC7575803
- DOI: 10.1016/j.redox.2020.101761
Stat2-Drp1 mediated mitochondrial mass increase is necessary for pro-inflammatory differentiation of macrophages
Erratum in
-
Corrigendum to 'Stat2-Drp1 mediated mitochondrial mass increase is necessary for pro-inflammatory differentiation of macrophages'[Redox Biology 37 (2020) 101761].Redox Biol. 2021 Feb;39:101786. doi: 10.1016/j.redox.2020.101786. Epub 2020 Dec 29. Redox Biol. 2021. PMID: 33384255 Free PMC article. No abstract available.
Abstract
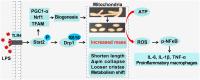
Macrophage recruitment and pro-inflammatory differentiation are hallmarks of various diseases, including infection and sepsis. Although studies suggest that mitochondria may regulate macrophage immune responses, it remains unclear whether mitochondrial mass affects macrophage pro-inflammatory differentiation. Here, we found that lipopolysaccharide (LPS)-activated macrophages possess higher mitochondrial mass than resting cells. Therefore, this study aimed to explore the functional role and molecular mechanisms of increased mitochondrial mass in pro-inflammatory differentiated macrophages. Results show that an increase in the mitochondrial mass of macrophages positively correlates with inflammatory cytokine generation in response to LPS. RNA-seq analysis revealed that LPS promotes signal transducers and activators of transcription 2 (Stat2) and dynamin-related protein 1 (Drp1) expression, which are enriched in positive mitochondrial fission regulation. Meanwhile, knockdown or pharmacological inhibition of Drp1 blunts LPS-induced mitochondrial mass increase and pro-inflammatory differentiation. Moreover, Stat2 boosts Drp1 phosphorylation at serine 616, required for Drp1-mediated mitochondrial fission. LPS also causes Stat2-and Drp1-dependent biogenesis, which contributes to the generation of additional mitochondria. However, these mitochondria are profoundly remodeled, displaying fragmented morphology, loose cristae, reduced Δψm, and metabolic programming. Furthermore, these remodeled mitochondria shift their function from ATP synthesis to reactive oxygen species (ROS) production, which drives NFκB-dependent inflammatory cytokine transcription. Interestingly, an increase in mitochondrial mass with constitutively active phosphomimetic mutant of Drp1 (Drp1S616E) boosted pro-inflammatory response in macrophages without LPS stimulation. In vivo, we also demonstrated that Mdivi-1 administration inhibits LPS-induced macrophage pro-inflammatory differentiation. Importantly, we observed Stat2 phosphorylation and Drp1-dependent mitochondrial mass increase in macrophages isolated from LPS-challenged mice. In conclusion, we comprehensively demonstrate that a Stat2-Drp1 dependent mitochondrial mass increase is necessary for pro-inflammatory differentiation of macrophages. Therefore, targeting the Stat2-Drp1 axis may provide novel therapeutic approaches for treating infection and inflammatory diseases.
Keywords: Drp1; Lipopolysaccharide; Mitochondrial mass; Pro-inflammatory macrophage; Reactive oxygen species; Stat2.
Copyright © 2020 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





References
-
- Bashir S., Sharma Y., Elahi A. Macrophage polarization: the link between inflammation and related diseases. Inflamm. Res. 2016;65(1):1–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

