Crucial Role of the C-Terminal Domain of Hfq Protein in Genomic Instability
- PMID: 33080799
- PMCID: PMC7603069
- DOI: 10.3390/microorganisms8101598
Crucial Role of the C-Terminal Domain of Hfq Protein in Genomic Instability
Abstract
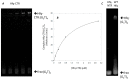
G-rich DNA repeats that can form G-quadruplex structures are prevalent in bacterial genomes and are frequently associated with regulatory regions of genes involved in virulence, antigenic variation, and antibiotic resistance. These sequences are also inherently mutagenic and can lead to changes affecting cell survival and adaptation. Transcription of the G-quadruplex-forming repeat (G3T)n in E. coli, when mRNA comprised the G-rich strand, promotes G-quadruplex formation in DNA and increases rates of deletion of G-quadruplex-forming sequences. The genomic instability of G-quadruplex repeats may be a source of genetic variability that can influence alterations and evolution of bacteria. The DNA chaperone Hfq is involved in the genetic instability of these G-quadruplex sequences. Inactivation of the hfq gene decreases the genetic instability of G-quadruplex, demonstrating that the genomic instability of this regulatory element can be influenced by the E. coli highly pleiotropic Hfq protein, which is involved in small noncoding RNA regulation pathways, and DNA organization and packaging. We have shown previously that the protein binds to and stabilizes these sequences, increasing rates of their genomic instability. Here, we extend this analysis to characterize the role of the C-terminal domain of Hfq protein in interaction with G-quadruplex structures. This allows to better understand the function of this specific region of the Hfq protein in genomic instability.
Keywords: DNA-directed mutagenesis; bacterial chromatin; genomic instability; nucleoid; quadruplex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Sinden R.R. DNA Structure and Function. Academic Press; Cambridge, MA, USA: 1994.
Grants and funding
LinkOut - more resources
Full Text Sources

