Three-Dimensional Structures of Carbohydrates and Where to Find Them
- PMID: 33081008
- PMCID: PMC7593929
- DOI: 10.3390/ijms21207702
Three-Dimensional Structures of Carbohydrates and Where to Find Them
Abstract
Analysis and systematization of accumulated data on carbohydrate structural diversity is a subject of great interest for structural glycobiology. Despite being a challenging task, development of computational methods for efficient treatment and management of spatial (3D) structural features of carbohydrates breaks new ground in modern glycoscience. This review is dedicated to approaches of chemo- and glyco-informatics towards 3D structural data generation, deposition and processing in regard to carbohydrates and their derivatives. Databases, molecular modeling and experimental data validation services, and structure visualization facilities developed for last five years are reviewed.
Keywords: PDB glycans; carbohydrate; database; glycoinformatics; model build; molecular modeling; spatial structure; structure validation; structure visualization; web-tool.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
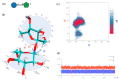
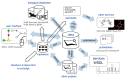
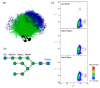
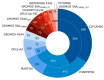


Similar articles
-
Bioinformatics and molecular modeling in glycobiology.Cell Mol Life Sci. 2010 Aug;67(16):2749-72. doi: 10.1007/s00018-010-0352-4. Epub 2010 Apr 4. Cell Mol Life Sci. 2010. PMID: 20364395 Free PMC article. Review.
-
3DSDSCAR--a three dimensional structural database for sialic acid-containing carbohydrates through molecular dynamics simulation.Carbohydr Res. 2010 Sep 23;345(14):2030-7. doi: 10.1016/j.carres.2010.06.021. Epub 2010 Jul 8. Carbohydr Res. 2010. PMID: 20691432
-
Data mining the PDB for glyco-related data.Methods Mol Biol. 2009;534:293-310. doi: 10.1007/978-1-59745-022-5_21. Methods Mol Biol. 2009. PMID: 19277543 Review.
-
Tools to assist determination and validation of carbohydrate 3D structure data.Methods Mol Biol. 2015;1273:229-40. doi: 10.1007/978-1-4939-2343-4_17. Methods Mol Biol. 2015. PMID: 25753715
-
CHARMM-GUI Glycan Modeler for modeling and simulation of carbohydrates and glycoconjugates.Glycobiology. 2019 Apr 1;29(4):320-331. doi: 10.1093/glycob/cwz003. Glycobiology. 2019. PMID: 30689864 Free PMC article.
Cited by
-
Complex Carbohydrates and Glycoconjugates: Structure, Functions and Applications.Int J Mol Sci. 2021 Nov 12;22(22):12219. doi: 10.3390/ijms222212219. Int J Mol Sci. 2021. PMID: 34830101 Free PMC article.
-
Current status of PTMs structural databases: applications, limitations and prospects.Amino Acids. 2022 Apr;54(4):575-590. doi: 10.1007/s00726-021-03119-z. Epub 2022 Jan 12. Amino Acids. 2022. PMID: 35020020 Review.
-
MonteCarbo: A software to generate and dock multifunctionalized ring molecules.J Comput Chem. 2021 Aug 5;42(21):1526-1534. doi: 10.1002/jcc.26559. Epub 2021 May 13. J Comput Chem. 2021. PMID: 33982793 Free PMC article.
-
Carbohydrate Structure Database: current state and recent developments.Anal Bioanal Chem. 2025 Feb;417(5):1025-1034. doi: 10.1007/s00216-024-05383-w. Epub 2024 Jun 25. Anal Bioanal Chem. 2025. PMID: 38914734 Review.
-
Glycan Shape, Motions, and Interactions Explored by NMR Spectroscopy.JACS Au. 2024 Jan 3;4(1):20-39. doi: 10.1021/jacsau.3c00639. eCollection 2024 Jan 22. JACS Au. 2024. PMID: 38274261 Free PMC article. Review.
References
-
- Buddhadeb M., Dimitrios M. Applications of Molecular Dynamics Simulations in Immunology: A Useful Computational Method in Aiding Vaccine Design. Curr. Proteom. 2006;3:259–270.
-
- Kuttel M.M., Ravenscroft N. Carbohydrate-Based Vaccines: From Concept to Clinic. Volume 1290. American Chemical Society; Washington, DC, USA: 2018. The Role of Molecular Modeling in Predicting Carbohydrate Antigen Conformation and Understanding Vaccine Immunogenicity; pp. 139–173.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

