Three-Dimensional Structures of Carbohydrates and Where to Find Them
- PMID: 33081008
- PMCID: PMC7593929
- DOI: 10.3390/ijms21207702
Three-Dimensional Structures of Carbohydrates and Where to Find Them
Abstract
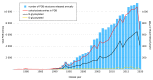
Analysis and systematization of accumulated data on carbohydrate structural diversity is a subject of great interest for structural glycobiology. Despite being a challenging task, development of computational methods for efficient treatment and management of spatial (3D) structural features of carbohydrates breaks new ground in modern glycoscience. This review is dedicated to approaches of chemo- and glyco-informatics towards 3D structural data generation, deposition and processing in regard to carbohydrates and their derivatives. Databases, molecular modeling and experimental data validation services, and structure visualization facilities developed for last five years are reviewed.
Keywords: PDB glycans; carbohydrate; database; glycoinformatics; model build; molecular modeling; spatial structure; structure validation; structure visualization; web-tool.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
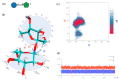
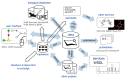
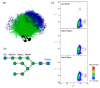
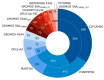
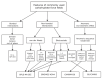
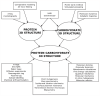
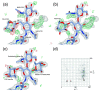

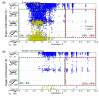

References
-
- Buddhadeb M., Dimitrios M. Applications of Molecular Dynamics Simulations in Immunology: A Useful Computational Method in Aiding Vaccine Design. Curr. Proteom. 2006;3:259–270.
-
- Kuttel M.M., Ravenscroft N. Carbohydrate-Based Vaccines: From Concept to Clinic. Volume 1290. American Chemical Society; Washington, DC, USA: 2018. The Role of Molecular Modeling in Predicting Carbohydrate Antigen Conformation and Understanding Vaccine Immunogenicity; pp. 139–173.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

