Diversity of tRNA Clusters in the Chloroviruses
- PMID: 33081353
- PMCID: PMC7589089
- DOI: 10.3390/v12101173
Diversity of tRNA Clusters in the Chloroviruses
Abstract
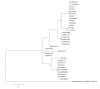
Viruses rely on their host's translation machinery for the synthesis of their own proteins. Problems belie viral translation when the host has a codon usage bias (CUB) that is different from an infecting virus due to differences in the GC content between the host and virus genomes. Here, we examine the hypothesis that chloroviruses adapted to host CUB by acquisition and selection of tRNAs that at least partially favor their own CUB. The genomes of 41 chloroviruses comprising three clades, each infecting a different algal host, have been sequenced, assembled and annotated. All 41 viruses not only encode tRNAs, but their tRNA genes are located in clusters. While differences were observed between clades and even within clades, seven tRNA genes were common to all three clades of chloroviruses, including the tRNAArg gene, which was found in all 41 chloroviruses. By comparing the codon usage of one chlorovirus algal host, in which the genome has been sequenced and annotated (67% GC content), to that of two of its viruses (40% GC content), we found that the viruses were able to at least partially overcome the host's CUB by encoding tRNAs that recognize AU-rich codons. Evidence presented herein supports the hypothesis that a chlorovirus tRNA cluster was present in the most recent common ancestor (MRCA) prior to divergence into three clades. In addition, the MRCA encoded a putative isoleucine lysidine synthase (TilS) that remains in 39/41 chloroviruses examined herein, suggesting a strong evolutionary pressure to retain the gene. TilS alters the anticodon of tRNAMet that normally recognizes AUG to then recognize AUA, a codon for isoleucine. This is advantageous to the chloroviruses because the AUA codon is 12-13 times more common in the chloroviruses than their host, further helping the chloroviruses to overcome CUB. Among large DNA viruses infecting eukaryotes, the presence of tRNA genes and tRNA clusters appear to be most common in the Phycodnaviridae and, to a lesser extent, in the Mimiviridae.
Keywords: algal viruses; chloroviruses; codon usage bias (CUB); tRNA clusters; tRNAs.
Conflict of interest statement
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
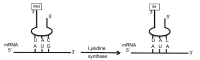
References
-
- Santini S., Jeudy S., Bartoli J., Poirot O., Lescot M., Abergel C., Barbe V., Wommack K.E., Noordeloos A.A.M., Brussaard C.P.D., et al. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc. Natl. Acad. Sci. USA. 2013;110:10800–10805. doi: 10.1073/pnas.1303251110. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

