Comparative Study of Pine Reference Genomes Reveals Transposable Element Interconnected Gene Networks
- PMID: 33081418
- PMCID: PMC7602945
- DOI: 10.3390/genes11101216
Comparative Study of Pine Reference Genomes Reveals Transposable Element Interconnected Gene Networks
Abstract
Sequencing the giga-genomes of several pine species has enabled comparative genomic analyses of these outcrossing tree species. Previous studies have revealed the wide distribution and extraordinary diversity of transposable elements (TEs) that occupy the large intergenic spaces in conifer genomes. In this study, we analyzed the distribution of TEs in gene regions of the assembled genomes of Pinus taeda and Pinus lambertiana using high-performance computing resources. The quality of draft genomes and the genome annotation have significant consequences for the investigation of TEs and these aspects are discussed. Several TE families frequently inserted into genes or their flanks were identified in both species' genomes. Potentially important sequence motifs were identified in TEs that could bind additional regulatory factors, promoting gene network formation with faster or enhanced transcription initiation. Node genes that contain many TEs were observed in multiple potential transposable element-associated networks. This study demonstrated the increased accumulation of TEs in the introns of stress-responsive genes of pines and suggests the possibility of rewiring them into responsive networks and sub-networks interconnected with node genes containing multiple TEs. Many such regulatory influences could lead to the adaptive environmental response clines that are characteristic of naturally spread pine populations.
Keywords: MITE; Pinus lambertiana; Pinus taeda; gene networks; gene regulation; introns; node gene; pine reference genome; retrotransposons; transposable elements.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

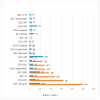

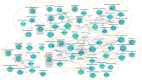
Similar articles
-
The Chinese pine genome and methylome unveil key features of conifer evolution.Cell. 2022 Jan 6;185(1):204-217.e14. doi: 10.1016/j.cell.2021.12.006. Epub 2021 Dec 28. Cell. 2022. PMID: 34965378
-
Sequence of the Sugar Pine Megagenome.Genetics. 2016 Dec;204(4):1613-1626. doi: 10.1534/genetics.116.193227. Epub 2016 Oct 28. Genetics. 2016. PMID: 27794028 Free PMC article.
-
Nearby transposable elements impact plant stress gene regulatory networks: a meta-analysis in A. thaliana and S. lycopersicum.BMC Genomics. 2022 Jan 4;23(1):18. doi: 10.1186/s12864-021-08215-8. BMC Genomics. 2022. PMID: 34983397 Free PMC article.
-
Mobility connects: transposable elements wire new transcriptional networks by transferring transcription factor binding motifs.Biochem Soc Trans. 2020 Jun 30;48(3):1005-1017. doi: 10.1042/BST20190937. Biochem Soc Trans. 2020. PMID: 32573687 Free PMC article. Review.
-
Transposable elements as genetic accelerators of evolution: contribution to genome size, gene regulatory network rewiring and morphological innovation.Genes Genet Syst. 2020 Jan 30;94(6):269-281. doi: 10.1266/ggs.19-00029. Epub 2020 Jan 10. Genes Genet Syst. 2020. PMID: 31932541 Review.
Cited by
-
Retrotransposons: How the continuous evolutionary front shapes plant genomes for response to heat stress.Front Plant Sci. 2022 Dec 9;13:1064847. doi: 10.3389/fpls.2022.1064847. eCollection 2022. Front Plant Sci. 2022. PMID: 36570931 Free PMC article. Review.
-
Distinct composition and amplification dynamics of transposable elements in sacred lotus (Nelumbo nucifera Gaertn.).Plant J. 2022 Oct;112(1):172-192. doi: 10.1111/tpj.15938. Epub 2022 Aug 26. Plant J. 2022. PMID: 35959634 Free PMC article.
-
DNA profiling and assessment of genetic diversity of relict species Allium altaicum Pall. on the territory of Altai.PeerJ. 2021 Jan 8;9:e10674. doi: 10.7717/peerj.10674. eCollection 2021. PeerJ. 2021. PMID: 33510974 Free PMC article.
-
Differential microRNA and Target Gene Expression in Scots Pine (Pinus sylvestris L.) Needles in Response to Methyl Jasmonate Treatment.Genes (Basel). 2024 Dec 27;16(1):26. doi: 10.3390/genes16010026. Genes (Basel). 2024. PMID: 39858573 Free PMC article.
References
-
- Zhou H., Liu Q., Li J., Jiang D., Zhou L., Wu P., Lu S., Li F., Zhu L., Liu Z., et al. Photoperiod-and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res. 2012;22:649–660. doi: 10.1038/cr.2012.28. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

