Evaluation of the Autof MS1000 mass spectrometer in the identification of clinical isolates
- PMID: 33081722
- PMCID: PMC7576717
- DOI: 10.1186/s12866-020-02005-0
Evaluation of the Autof MS1000 mass spectrometer in the identification of clinical isolates
Abstract
Background: To evaluate the accuracy and performance of the Autof MS1000 mass spectrometer in bacteria and yeast identification, 2342 isolates were obtained from microbial cultures of clinical specimens (e.g. blood, cerebrospinal fluid, respiratory tract samples, lumbar puncture fluid, wound samples, stool, and urine) collected in 2019 in Henan Provincial People's Hospital. Repetitive strains from the same patient were excluded. We tested the Autof MS1000 and Bruker Biotyper mass spectrometry systems and the classical biochemical identification system VITEK 2/API 20C AUX. Inconsistencies in strain identification among the three systems were identified by 16S rDNA and gene sequencing.
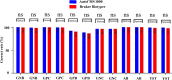
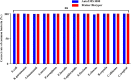
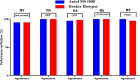
Results: At the species level, the Autof MS1000 and Bruker Biotyper systems had isolate identification accuracies of 98.9 and 98.5%, respectively. At the genus level, the Autof MS1000 and Bruker Biotyper systems were 99.7 and 99.4% accurate, respectively. The instruments did not significantly differ in identification accuracy at either taxonomic level. The frequencies of unreliable identification were 1.1% (26/2342) for the Autof MS1000 and 1.5% (34/2342) for the Bruker Biotyper. In vitro experiments demonstrated that the coincidence rate of the Autof MS1000 mass spectrometer in the identification of five types of bacteria was > 93%, the identification error rate was < 3%, and the no identification rate was 0. This indicates that the Autof MS1000 system is acceptable for identification.
Conclusions: The Autof MS1000 mass spectrometer can be utilised to identify clinical isolates. However, an upgradation of the database is recommended to correctly identify rare strains.
Keywords: Autof MS1000; Bacterial identification; Bruker Biotyper; Clinical samples; Mass spectrometry; Performance verification.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Martiny D, Busson L, Wybo I, El Haj RA, Dediste A, Vandenberg O. Comparison of the microflex LT and Vitek MS systems for the routine identification of bacteria by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry. J Clin Microbiol. 2012;50:1313–1325. doi: 10.1128/JCM.05971-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

