Bioengineered human skeletal muscle capable of functional regeneration
- PMID: 33081771
- PMCID: PMC7576716
- DOI: 10.1186/s12915-020-00884-3
Bioengineered human skeletal muscle capable of functional regeneration
Abstract
Background: Skeletal muscle (SkM) regenerates following injury, replacing damaged tissue with high fidelity. However, in serious injuries, non-regenerative defects leave patients with loss of function, increased re-injury risk and often chronic pain. Progress in treating these non-regenerative defects has been slow, with advances only occurring where a comprehensive understanding of regeneration has been gained. Tissue engineering has allowed the development of bioengineered models of SkM which regenerate following injury to support research in regenerative physiology. To date, however, no studies have utilised human myogenic precursor cells (hMPCs) to closely mimic functional human regenerative physiology.
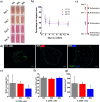
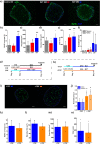
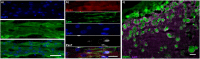
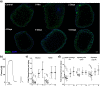
Results: Here we address some of the difficulties associated with cell number and hMPC mitogenicity using magnetic association cell sorting (MACS), for the marker CD56, and media supplementation with fibroblast growth factor 2 (FGF-2) and B-27 supplement. Cell sorting allowed extended expansion of myogenic cells and supplementation was shown to improve myogenesis within engineered tissues and force generation at maturity. In addition, these engineered human SkM regenerated following barium chloride (BaCl2) injury. Following injury, reductions in function (87.5%) and myotube number (33.3%) were observed, followed by a proliferative phase with increased MyoD+ cells and a subsequent recovery of function and myotube number. An expansion of the Pax7+ cell population was observed across recovery suggesting an ability to generate Pax7+ cells within the tissue, similar to the self-renewal of satellite cells seen in vivo.
Conclusions: This work outlines an engineered human SkM capable of functional regeneration following injury, built upon an open source system adding to the pre-clinical testing toolbox to improve the understanding of basic regenerative physiology.
Keywords: Regeneration; Satellite cell; Skeletal muscle; Tissue engineering.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

