SNP barcodes provide higher resolution than microsatellite markers to measure Plasmodium vivax population genetics
- PMID: 33081815
- PMCID: PMC7576724
- DOI: 10.1186/s12936-020-03440-0
SNP barcodes provide higher resolution than microsatellite markers to measure Plasmodium vivax population genetics
Abstract
Background: Genomic surveillance of malaria parasite populations has the potential to inform control strategies and to monitor the impact of interventions. Barcodes comprising large numbers of single nucleotide polymorphism (SNP) markers are accurate and efficient genotyping tools, however may need to be tailored to specific malaria transmission settings, since 'universal' barcodes can lack resolution at the local scale. A SNP barcode was developed that captures the diversity and structure of Plasmodium vivax populations of Papua New Guinea (PNG) for research and surveillance.
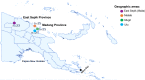
Methods: Using 20 high-quality P. vivax genome sequences from PNG, a total of 178 evenly spaced neutral SNPs were selected for development of an amplicon sequencing assay combining a series of multiplex PCRs and sequencing on the Illumina MiSeq platform. For initial testing, 20 SNPs were amplified in a small number of mono- and polyclonal P. vivax infections. The full barcode was then validated by genotyping and population genetic analyses of 94 P. vivax isolates collected between 2012 and 2014 from four distinct catchment areas on the highly endemic north coast of PNG. Diversity and population structure determined from the SNP barcode data was then benchmarked against that of ten microsatellite markers used in previous population genetics studies.
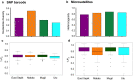
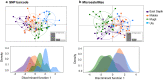
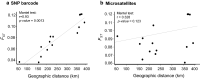
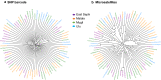
Results: From a total of 28,934,460 reads generated from the MiSeq Illumina run, 87% mapped to the PvSalI reference genome with deep coverage (median = 563, range 56-7586) per locus across genotyped samples. Of 178 SNPs assayed, 146 produced high-quality genotypes (minimum coverage = 56X) in more than 85% of P. vivax isolates. No amplification bias was introduced due to either polyclonal infection or whole genome amplification (WGA) of samples before genotyping. Compared to the microsatellite panels, the SNP barcode revealed greater variability in genetic diversity between populations and geographical population structure. The SNP barcode also enabled assignment of genotypes according to their geographic origins with a significant association between genetic distance and geographic distance at the sub-provincial level.
Conclusions: High-throughput SNP barcoding can be used to map variation of malaria transmission dynamics at sub-national resolution. The low cost per sample and genotyping strategy makes the transfer of this technology to field settings highly feasible.
Keywords: Diversity; Malaria; Microsatellites; Papua New Guinea; Plasmodium vivax; Population structure; Single Nucleotide Polymorphisms (SNPs).
Conflict of interest statement
The authors declare that they have no any competing interests.
Figures
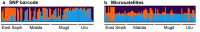
References
-
- WHO. World malaria report 2019. Geneva: World Health Organization; 2019.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

