Incipient sympatric speciation in wild barley caused by geological-edaphic divergence
- PMID: 33082129
- PMCID: PMC7652381
- DOI: 10.26508/lsa.202000827
Incipient sympatric speciation in wild barley caused by geological-edaphic divergence
Abstract
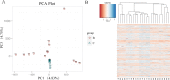
Sympatric speciation (SS) has been contentious since the idea was suggested by Darwin. Here, we show in wild barley SS due to geologic and edaphic divergence in "Evolution Plateau," Upper Galilee, Israel. Our whole genome resequencing data showed SS separating between the progenitor old Senonian chalk and abutting derivative young Pleistocene basalt wild barley populations. The basalt wild barley species unfolds larger effective population size, lower recombination rates, and larger genetic diversity. Both species populations show similar descending trend ∼200,000 yr ago associated with the last glacial maximum. Coalescent demography analysis indicates that SS was local, primary, in situ, and not due to a secondary contact from ex situ allopatric population. Adaptive divergent putatively selected genes were identified in both populations. Remarkably, disease resistant genes were selected in the wet basalt population, and genes related to flowering time, leading to temporal reproductive isolation, were selected in the chalk population. The evidence substantiates adaptive ecological SS in wild barley, highlighting the genome landscape during SS with gene flow, due to geologic-edaphic divergence.
© 2020 Li et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
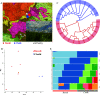


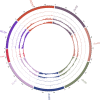

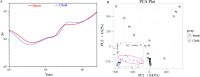


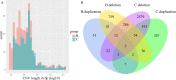


References
-
- Bolnick DI, Fitzpatrick BM (2007) Sympatric speciation: Models and empirical evidence. Annu Rev Ecol Evol Syst 38: 459–487. 10.1146/annurev.ecolsys.38.091206.095804 - DOI
-
- Brown A, Nevo E, Zohary D, Dagan O (1978) Genetic variation in natural populations of wild barley (Hordeum spontaneum). Genetica 49: 97–108. 10.1007/bf00120555 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
