Inhibition of Coronavirus Entry In Vitro and Ex Vivo by a Lipid-Conjugated Peptide Derived from the SARS-CoV-2 Spike Glycoprotein HRC Domain
- PMID: 33082259
- PMCID: PMC7587434
- DOI: 10.1128/mBio.01935-20
Inhibition of Coronavirus Entry In Vitro and Ex Vivo by a Lipid-Conjugated Peptide Derived from the SARS-CoV-2 Spike Glycoprotein HRC Domain
Abstract
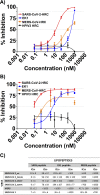
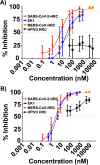
The emergence of severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), the etiological agent of the 2019 coronavirus disease (COVID-19), has erupted into a global pandemic that has led to tens of millions of infections and hundreds of thousands of deaths worldwide. The development of therapeutics to treat infection or as prophylactics to halt viral transmission and spread is urgently needed. SARS-CoV-2 relies on structural rearrangements within a spike (S) glycoprotein to mediate fusion of the viral and host cell membranes. Here, we describe the development of a lipopeptide that is derived from the C-terminal heptad repeat (HRC) domain of SARS-CoV-2 S that potently inhibits infection by SARS-CoV-2. The lipopeptide inhibits cell-cell fusion mediated by SARS-CoV-2 S and blocks infection by live SARS-CoV-2 in Vero E6 cell monolayers more effectively than previously described lipopeptides. The SARS-CoV-2 lipopeptide exhibits broad-spectrum activity by inhibiting cell-cell fusion mediated by SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV) and blocking infection by live MERS-CoV in cell monolayers. We also show that the SARS-CoV-2 HRC-derived lipopeptide potently blocks the spread of SARS-CoV-2 in human airway epithelial (HAE) cultures, an ex vivo model designed to mimic respiratory viral propagation in humans. While viral spread of SARS-CoV-2 infection was widespread in untreated airways, those treated with SARS-CoV-2 HRC lipopeptide showed no detectable evidence of viral spread. These data provide a framework for the development of peptide therapeutics for the treatment of or prophylaxis against SARS-CoV-2 as well as other coronaviruses.IMPORTANCE SARS-CoV-2, the causative agent of COVID-19, continues to spread globally, placing strain on health care systems and resulting in rapidly increasing numbers of cases and mortalities. Despite the growing need for medical intervention, no FDA-approved vaccines are yet available, and treatment has been limited to supportive therapy for the alleviation of symptoms. Entry inhibitors could fill the important role of preventing initial infection and preventing spread. Here, we describe the design, synthesis, and evaluation of a lipopeptide that is derived from the HRC domain of the SARS-CoV-2 S glycoprotein that potently inhibits fusion mediated by SARS-CoV-2 S glycoprotein and blocks infection by live SARS-CoV-2 in both cell monolayers (in vitro) and human airway tissues (ex vivo). Our results highlight the SARS-CoV-2 HRC-derived lipopeptide as a promising therapeutic candidate for SARS-CoV-2 infections.
Keywords: SARS-CoV-2; fusion inhibitor; lipopeptide; spike protein.
Copyright © 2020 Outlaw et al.
Figures


References
-
- Corbett KS, Edwards D, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, Himansu S, Schäfer A, Ziwawo CT, DiPiazza AT, Dinnon KH, Elbashir SM, Shaw CA, Woods A, Fritch EJ, Martinez DR, Bock KW, Minai M, Nagata BM, Hutchinson GB, Bahl K, Garcia-Dominguez D, Ma L, Renzi I, Kong W-P, Schmidt SD, Wang L, Zhang Y, Stevens LJ, Phung E, Chang LA, Loomis RJ, Altaras NE, Narayanan E, Metkar M, Presnyak V, Liu C, Louder MK, Shi W, Leung K, Yang ES, West A, Gully KL, Wang N, Wrapp D, Doria-Rose NA, Stewart-Jones G, Bennett H, Nason MC, Ruckwardt TJ, McLellan JS, et al. 2020. SARS-CoV-2 mRNA vaccine development enabled by prototype pathogen preparedness. bioRxiv doi: 10.1101/2020.06.11.145920. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
