Chemical-Genetic Interactions with the Proline Analog L-Azetidine-2-Carboxylic Acid in Saccharomyces cerevisiae
- PMID: 33082270
- PMCID: PMC7718759
- DOI: 10.1534/g3.120.401876
Chemical-Genetic Interactions with the Proline Analog L-Azetidine-2-Carboxylic Acid in Saccharomyces cerevisiae
Abstract
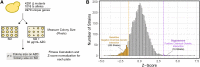
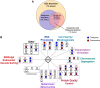
Non-proteinogenic amino acids, such as the proline analog L-azetidine-2-carboxylic acid (AZC), are detrimental to cells because they are mis-incorporated into proteins and lead to proteotoxic stress. Our goal was to identify genes that show chemical-genetic interactions with AZC in Saccharomyces cerevisiae and thus also potentially define the pathways cells use to cope with amino acid mis-incorporation. Screening the yeast deletion and temperature sensitive collections, we found 72 alleles with negative chemical-genetic interactions with AZC treatment and 12 alleles that suppress AZC toxicity. Many of the genes with negative chemical-genetic interactions are involved in protein quality control pathways through the proteasome. Genes involved in actin cytoskeleton organization and endocytosis also had negative chemical-genetic interactions with AZC. Related to this, the number of actin patches per cell increases upon AZC treatment. Many of the same cellular processes were identified to have interactions with proteotoxic stress caused by two other amino acid analogs, canavanine and thialysine, or a mistranslating tRNA variant that mis-incorporates serine at proline codons. Alleles that suppressed AZC-induced toxicity functioned through the amino acid sensing TOR pathway or controlled amino acid permeases required for AZC uptake. Further suggesting the potential of genetic changes to influence the cellular response to proteotoxic stress, overexpressing many of the genes that had a negative chemical-genetic interaction with AZC suppressed AZC toxicity.
Keywords: L-azetidine-2-carboxylic acid; Saccharomyces cerevisiae; actin cytoskeleton; mistranslation; protein quality control; proteotoxic stress.
Copyright © 2020 Berg et al.
Figures





Similar articles
-
Involvement of the stress-responsive transcription factor gene MSN2 in the control of amino acid uptake in Saccharomyces cerevisiae.FEMS Yeast Res. 2019 Aug 1;19(5):foz052. doi: 10.1093/femsyr/foz052. FEMS Yeast Res. 2019. PMID: 31328231
-
TORC1 regulates autophagy induction in response to proteotoxic stress in yeast and human cells.Biochem Biophys Res Commun. 2019 Apr 2;511(2):434-439. doi: 10.1016/j.bbrc.2019.02.077. Epub 2019 Feb 21. Biochem Biophys Res Commun. 2019. PMID: 30797551
-
A novel acetyltransferase found in Saccharomyces cerevisiae Sigma1278b that detoxifies a proline analogue, azetidine-2-carboxylic acid.J Biol Chem. 2001 Nov 9;276(45):41998-2002. doi: 10.1074/jbc.C100487200. Epub 2001 Sep 12. J Biol Chem. 2001. PMID: 11555637
-
Properties, metabolisms, and applications of (L)-proline analogues.Appl Microbiol Biotechnol. 2013 Aug;97(15):6623-34. doi: 10.1007/s00253-013-5022-7. Epub 2013 Jun 19. Appl Microbiol Biotechnol. 2013. PMID: 23780584 Review.
-
A comprehensive review of the proline mimic azetidine-2-carboxylic acid (A2C).Toxicology. 2025 Jan;510:153999. doi: 10.1016/j.tox.2024.153999. Epub 2024 Nov 15. Toxicology. 2025. PMID: 39549916 Review.
Cited by
-
A novel mistranslating tRNA model in Drosophila melanogaster has diverse, sexually dimorphic effects.G3 (Bethesda). 2022 May 6;12(5):jkac035. doi: 10.1093/g3journal/jkac035. G3 (Bethesda). 2022. PMID: 35143655 Free PMC article.
-
URA6 mutations provide an alternative mechanism for 5-FOA resistance in Saccharomyces cerevisiae.bioRxiv [Preprint]. 2024 Jun 5:2024.06.03.597250. doi: 10.1101/2024.06.03.597250. bioRxiv. 2024. PMID: 38895202 Free PMC article. Preprint.
-
Genetic inactivation of essential HSF1 reveals an isolated transcriptional stress response selectively induced by protein misfolding.Mol Biol Cell. 2023 Sep 1;34(10):ar101. doi: 10.1091/mbc.E23-05-0153. Epub 2023 Jul 19. Mol Biol Cell. 2023. PMID: 37467033 Free PMC article.
-
The amino acid substitution affects cellular response to mistranslation.G3 (Bethesda). 2021 Sep 27;11(10):jkab218. doi: 10.1093/g3journal/jkab218. G3 (Bethesda). 2021. PMID: 34568909 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
