Neuropilin-1 is a host factor for SARS-CoV-2 infection
- PMID: 33082294
- PMCID: PMC7612957
- DOI: 10.1126/science.abd3072
Neuropilin-1 is a host factor for SARS-CoV-2 infection
Abstract
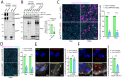
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), uses the viral spike (S) protein for host cell attachment and entry. The host protease furin cleaves the full-length precursor S glycoprotein into two associated polypeptides: S1 and S2. Cleavage of S generates a polybasic Arg-Arg-Ala-Arg carboxyl-terminal sequence on S1, which conforms to a C-end rule (CendR) motif that binds to cell surface neuropilin-1 (NRP1) and NRP2 receptors. We used x-ray crystallography and biochemical approaches to show that the S1 CendR motif directly bound NRP1. Blocking this interaction by RNA interference or selective inhibitors reduced SARS-CoV-2 entry and infectivity in cell culture. NRP1 thus serves as a host factor for SARS-CoV-2 infection and may potentially provide a therapeutic target for COVID-19.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
T. Teesalu is an inventor of patents on CendR peptides and shareholder of Cend Therapeutics Inc., a company that holds a license for the CendR peptides and is developing the peptides for cancer therapy. J. Hiscox is a member of the Department of Health, New and Emerging Respiratory Virus Threats Advisory Group (NERVTAG) and the Department of Health, Testing Advisory Group. U. Greber is a consultant to F. Hoffmann-La Roche Ltd, Switzerland. All other authors declare no competing interests.
Figures


Comment in
-
Enhancing host cell infection by SARS-CoV-2.Science. 2020 Nov 13;370(6518):765-766. doi: 10.1126/science.abf0732. Science. 2020. PMID: 33184193 No abstract available.
-
Not only ACE2-the quest for additional host cell mediators of SARS-CoV-2 infection: Neuropilin-1 (NRP1) as a novel SARS-CoV-2 host cell entry mediator implicated in COVID-19.Signal Transduct Target Ther. 2021 Jan 18;6(1):21. doi: 10.1038/s41392-020-00460-9. Signal Transduct Target Ther. 2021. PMID: 33462185 Free PMC article. No abstract available.
References
-
- WHO Coronavirus disease 2019 (COVID-19) Weekly Epidemiological. Update – 31 August 2020, https://www.who.int/docs/default-source/coronaviruse/situation-reports/2....
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

