Porcine circovirus 2 (PCV2) population study in experimentally infected pigs developing PCV2-systemic disease or a subclinical infection
- PMID: 33082419
- PMCID: PMC7576782
- DOI: 10.1038/s41598-020-74627-3
Porcine circovirus 2 (PCV2) population study in experimentally infected pigs developing PCV2-systemic disease or a subclinical infection
Abstract
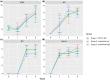
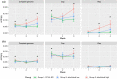
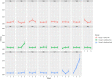
Porcine circovirus 2 (PCV2) is a single stranded DNA virus with one of the highest mutation rates among DNA viruses. This ability allows it to generate a cloud of mutants constantly providing new opportunities to adapt and evade the immune system. This pig pathogen is associated to many diseases, globally called porcine circovirus diseases (PCVD) and has been a threat to pig industry since its discovery in the early 90's. Although 11 ORFs have been predicted from its genome, only two main proteins have been deeply characterized, i.e. Rep and Cap. The structural Cap protein possesses the majority of the epitopic determinants of this non-enveloped virus. The evolution of PCV2 is affected by both natural and vaccine-induced immune responses, which enhances the genetic variability, especially in the most immunogenic Cap region. Intra-host variability has been also demonstrated in infected animals where long-lasting infections can take place. However, the association between this intra-host variability and pathogenesis has never been studied for this virus. Here, the within-host PCV2 variability was monitored over time by next generation sequencing during an experimental infection, demonstrating the presence of large heterogeneity. Remarkably, the level of quasispecies diversity, affecting particularly the Cap coding region, was statistically different depending on viremia levels and clinical signs detected after infection. Moreover, we proved the existence of hyper mutant subjects harboring a remarkably higher number of genetic variants. Altogether, these results suggest an interaction between genetic diversity, host immune system and disease severity.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Markedly different immune responses and virus kinetics in littermates infected with porcine circovirus type 2 or porcine parvovirus type 1.Vet Immunol Immunopathol. 2017 Sep;191:51-59. doi: 10.1016/j.vetimm.2017.08.003. Epub 2017 Aug 9. Vet Immunol Immunopathol. 2017. PMID: 28895867
-
Establishment of a Rep' protein antibody detection method to distinguish natural infection with PCV2 from subunit vaccine immunization.J Med Microbiol. 2020 Sep;69(9):1183-1196. doi: 10.1099/jmm.0.001230. Epub 2020 Aug 19. J Med Microbiol. 2020. PMID: 32812860
-
Effect of an interferon-stimulated response element (ISRE) mutant of porcine circovirus type 2 (PCV2) on PCV2-induced pathological lesions in a porcine reproductive and respiratory syndrome virus (PRRSV) co-infection model.Vet Microbiol. 2011 Jan 10;147(1-2):49-58. doi: 10.1016/j.vetmic.2010.06.010. Epub 2010 Jun 19. Vet Microbiol. 2011. PMID: 20637549
-
Porcine circovirus type 2 (PCV2): pathogenesis and interaction with the immune system.Annu Rev Anim Biosci. 2013 Jan;1:43-64. doi: 10.1146/annurev-animal-031412-103720. Epub 2013 Jan 3. Annu Rev Anim Biosci. 2013. PMID: 25387012 Review.
-
Porcine Circovirus Type 2 (PCV2) Vaccines in the Context of Current Molecular Epidemiology.Viruses. 2017 May 6;9(5):99. doi: 10.3390/v9050099. Viruses. 2017. PMID: 28481275 Free PMC article. Review.
Cited by
-
Phylodynamic analysis of current Porcine circovirus 4 sequences: Does the porcine circoviruses evolutionary history repeat itself?Transbound Emerg Dis. 2022 Sep;69(5):e3363-e3369. doi: 10.1111/tbed.14638. Epub 2022 Jun 30. Transbound Emerg Dis. 2022. PMID: 35735227 Free PMC article.
-
When Everything Becomes Bigger: Big Data for Big Poultry Production.Animals (Basel). 2023 May 30;13(11):1804. doi: 10.3390/ani13111804. Animals (Basel). 2023. PMID: 37889739 Free PMC article. Review.
-
Rapid and Easy-Read Porcine Circovirus Type 4 Detection with CRISPR-Cas13a-Based Lateral Flow Strip.Microorganisms. 2023 Jan 31;11(2):354. doi: 10.3390/microorganisms11020354. Microorganisms. 2023. PMID: 36838319 Free PMC article.
-
One Dose of a Novel Vaccine Containing Two Genotypes of Porcine Circovirus (PCV2a and PCV2b) and Mycoplasma hyopneumoniae Conferred a Duration of Immunity of 23 Weeks.Vaccines (Basel). 2021 Jul 29;9(8):834. doi: 10.3390/vaccines9080834. Vaccines (Basel). 2021. PMID: 34451959 Free PMC article.
-
Exploring the Cause of Diarrhoea and Poor Growth in 8-11-Week-Old Pigs from an Australian Pig Herd Using Metagenomic Sequencing.Viruses. 2021 Aug 13;13(8):1608. doi: 10.3390/v13081608. Viruses. 2021. PMID: 34452472 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

