The crucial choice of reference genes: identification of miR-191-5p for normalization of miRNAs expression in bone marrow mesenchymal stromal cell and HS27a/HS5 cell lines
- PMID: 33082452
- PMCID: PMC7576785
- DOI: 10.1038/s41598-020-74685-7
The crucial choice of reference genes: identification of miR-191-5p for normalization of miRNAs expression in bone marrow mesenchymal stromal cell and HS27a/HS5 cell lines
Abstract
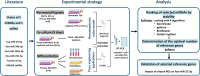
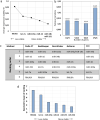
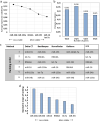
Bone marrow mesenchymal stromal cells (BM-MSCs) have a critical role in tissue regeneration and in the hematopoietic niche due to their differentiation and self-renewal capacities. These mechanisms are finely tuned partly by small non-coding microRNA implicated in post-transcriptional regulation. The easiest way to quantify them is RT-qPCR followed by normalization on validated reference genes (RGs). This study identified appropriate RG for normalization of miRNA expression in BM-MSCs and HS27a and HS5 cell lines in various conditions including normoxia, hypoxia, co-culture, as model for the hematopoietic niche and after induced differentiation as model for regenerative medicine. Six candidates, namely miR-16-5p, miR-34b-3p, miR-103a-3p, miR-191-5p, let-7a-5p and RNU6A were selected and their expression verified by RT-qPCR. Next, a ranking on stability of the RG candidates were performed with two algorithms geNorm and RefFinder and the optimal number of RGs needed to normalize was determined. Our results indicate miR-191-5p as the most stable miRNA in all conditions but also that RNU6a, usually used as RG is the less stable gene. This study demonstrates the interest of rigorously evaluating candidate miRNAs as reference genes and the importance of the normalization process to study the expression of miRNAs in BM-MSCs or derived cell lines.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

