The transcription factor Sox10 is an essential determinant of branching morphogenesis and involution in the mouse mammary gland
- PMID: 33082503
- PMCID: PMC7575560
- DOI: 10.1038/s41598-020-74664-y
The transcription factor Sox10 is an essential determinant of branching morphogenesis and involution in the mouse mammary gland
Abstract
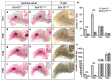
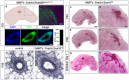
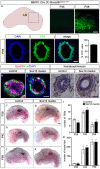
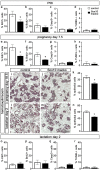
The high mobility group-domain containing transcription factor Sox10 is an essential regulator of developmental processes and homeostasis in the neural crest, several neural crest-derived lineages and myelinating glia. Recent studies have also implicated Sox10 as an important factor in mammary stem and precursor cells. Here we employ a series of mouse mutants with constitutive and conditional Sox10 deficiencies to show that Sox10 has multiple functions in the developing mammary gland. While there is no indication for a requirement of Sox10 in the specification of the mammary placode or descending mammary bud, it is essential for both the prenatal hormone-independent as well as the pubertal hormone-dependent branching of the mammary epithelium and for proper alveologenesis during pregnancy. It furthermore acts in a dosage-dependent manner. Sox10 also plays a role during the involution process at the end of the lactation period. Whereas its effect on epithelial branching and alveologenesis are likely causally related to its function in mammary stem and precursor cells, this is not the case for its function during involution where Sox10 seems to work at least in part through regulation of the miR-424(322)/503 cluster.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
HDAC1 and HDAC2 control the specification of neural crest cells into peripheral glia.J Neurosci. 2014 Apr 23;34(17):6112-22. doi: 10.1523/JNEUROSCI.5212-13.2014. J Neurosci. 2014. PMID: 24760871 Free PMC article.
-
Sox10-cre BAC transgenes reveal temporal restriction of mesenchymal cranial neural crest and identify glandular Sox10 expression.Dev Biol. 2021 Mar;471:119-137. doi: 10.1016/j.ydbio.2020.12.006. Epub 2020 Dec 13. Dev Biol. 2021. PMID: 33316258 Free PMC article.
-
SoxE proteins are differentially required in mouse adrenal gland development.Mol Biol Cell. 2008 Apr;19(4):1575-86. doi: 10.1091/mbc.e07-08-0782. Epub 2008 Feb 13. Mol Biol Cell. 2008. PMID: 18272785 Free PMC article.
-
The importance of having your SOX on: role of SOX10 in the development of neural crest-derived melanocytes and glia.Oncogene. 2003 May 19;22(20):3024-34. doi: 10.1038/sj.onc.1206442. Oncogene. 2003. PMID: 12789277 Review.
-
[Mammary gland development: Role of basal myoepithelial cells].J Soc Biol. 2006;200(2):193-8. doi: 10.1051/jbio:2006021. J Soc Biol. 2006. PMID: 17151555 Review. French.
Cited by
-
Embryonic Mammary Gland Morphogenesis.Adv Exp Med Biol. 2025;1464:9-27. doi: 10.1007/978-3-031-70875-6_2. Adv Exp Med Biol. 2025. PMID: 39821018 Review.
-
Association between Synonymous SNPs of SOX10 and Plumage Color and Reproductive Traits of Ducks.Animals (Basel). 2022 Nov 29;12(23):3345. doi: 10.3390/ani12233345. Animals (Basel). 2022. PMID: 36496864 Free PMC article.
-
Dynamic miRNA Landscape Links Mammary Gland Development to the Regulation of Milk Protein Expression in Mice.Animals (Basel). 2022 Mar 14;12(6):727. doi: 10.3390/ani12060727. Animals (Basel). 2022. PMID: 35327124 Free PMC article.
-
SOX factors as cell-state regulators in the mammary gland and breast cancer.Semin Cell Dev Biol. 2021 Jun;114:126-133. doi: 10.1016/j.semcdb.2021.01.002. Epub 2021 Feb 12. Semin Cell Dev Biol. 2021. PMID: 33583737 Free PMC article. Review.
-
Clinical Significance of SOX10 Expression in Human Pathology.Curr Issues Mol Biol. 2023 Dec 15;45(12):10131-10158. doi: 10.3390/cimb45120633. Curr Issues Mol Biol. 2023. PMID: 38132479 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

