Bi-paratopic and multivalent VH domains block ACE2 binding and neutralize SARS-CoV-2
- PMID: 33082574
- PMCID: PMC8356808
- DOI: 10.1038/s41589-020-00679-1
Bi-paratopic and multivalent VH domains block ACE2 binding and neutralize SARS-CoV-2
Abstract
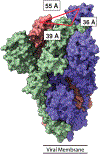
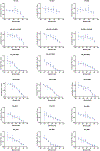
Neutralizing agents against SARS-CoV-2 are urgently needed for the treatment and prophylaxis of COVID-19. Here, we present a strategy to rapidly identify and assemble synthetic human variable heavy (VH) domains toward neutralizing epitopes. We constructed a VH-phage library and targeted the angiotensin-converting enzyme 2 (ACE2) binding interface of the SARS-CoV-2 Spike receptor-binding domain (Spike-RBD). Using a masked selection approach, we identified VH binders to two non-overlapping epitopes and further assembled these into multivalent and bi-paratopic formats. These VH constructs showed increased affinity to Spike (up to 600-fold) and neutralization potency (up to 1,400-fold) on pseudotyped SARS-CoV-2 virus when compared to standalone VH domains. The most potent binder, a trivalent VH, neutralized authentic SARS-CoV-2 with a half-maximal inhibitory concentration (IC50) of 4.0 nM (180 ng ml-1). A cryo-EM structure of the trivalent VH bound to Spike shows each VH domain engaging an RBD at the ACE2 binding site, confirming our original design strategy.
Conflict of interest statement
Competing Interests:
The authors declare no competing interests.
Figures


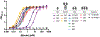







Update of
-
Bi-paratopic and multivalent human VH domains neutralize SARS-CoV-2 by targeting distinct epitopes within the ACE2 binding interface of Spike.bioRxiv [Preprint]. 2020 Aug 10:2020.08.08.242511. doi: 10.1101/2020.08.08.242511. bioRxiv. 2020. Update in: Nat Chem Biol. 2021 Jan;17(1):113-121. doi: 10.1038/s41589-020-00679-1. PMID: 32817948 Free PMC article. Updated. Preprint.
References
-
- Pinto D et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R35 GM122451/GM/NIGMS NIH HHS/United States
- F32 5F32CA239417/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)/International
- R35 GM122451-01/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)/International
- DRG-2204-14/Damon Runyon Cancer Research Foundation (Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation)/International
- F32 CA236151/CA/NCI NIH HHS/United States
- DRG-2297-17/Damon Runyon Cancer Research Foundation (Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation)/International
- HR0011-19-2-0020/United States Department of Defense | Defense Advanced Research Projects Agency (DARPA)/International
- T32 GM007618/GM/NIGMS NIH HHS/United States
- T32 HL007185/HL/NHLBI NIH HHS/United States
- F32 CA239417/CA/NCI NIH HHS/United States
- U19 AI135990/AI/NIAID NIH HHS/United States
- HL007185/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)/International
- 5F32CA236151-02/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

