The Histone Demethylase KDM3B Promotes Osteo-/Odontogenic Differentiation, Cell Proliferation, and Migration Potential of Stem Cells from the Apical Papilla
- PMID: 33082788
- PMCID: PMC7563049
- DOI: 10.1155/2020/8881021
The Histone Demethylase KDM3B Promotes Osteo-/Odontogenic Differentiation, Cell Proliferation, and Migration Potential of Stem Cells from the Apical Papilla
Abstract
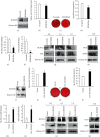
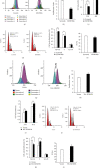
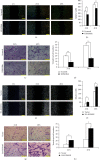
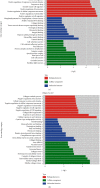
Understanding the regulation mechanisms of mesenchymal stem cells (MSCs) can assist in tissue regeneration. The histone demethylase (KDM) family has a crucial role in differentiation and cell proliferation of MSCs, while the function of KDM3B in MSCs is not well understood. In this study, we used the stem cells from the apical papilla (SCAPs) to test whether KDM3B could regulate the function of MSCs. By an alkaline phosphatase (ALP) activity assay, Alizarin red staining, real-time RT-PCR, and western blot analysis, we found that KDM3B enhanced the ALP activity and mineralization of SCAPs and promoted the expression of runt-related transcription factor 2 (RUNX2), osterix (OSX), dentin sialophosphoprotein (DSPP), and osteocalcin (OCN). Additionally, the CFSE, CCK-8, and flow cytometry assays revealed that KDM3B improved cell proliferation by accelerating cell cycle transition from the G1 to S phase. Scratch and transwell migration assays displayed that KDM3B promoted the migration potential of SCAPs. Mechanically, microarray results displayed that 98 genes were upregulated, including STAT1, CCND1, and FGF5, and 48 genes were downregulated after KDM3B overexpression. Besides, we found that the Toll-like receptor and JAK-STAT signaling pathway may be involved in the regulating function of KDM3B in SCAPs. In brief, we discovered that KDM3B promoted the osteo-/odontogenic differentiation, cell proliferation, and migration potential of SCAPs and provided a novel target and theoretical basis for regenerative medicine.
Copyright © 2020 Chen Zhang et al.
Conflict of interest statement
The authors declared that they have no competing interests.
Figures
Similar articles
-
Distal-less homeobox 5 promotes the osteo-/dentinogenic differentiation potential of stem cells from apical papilla by activating histone demethylase KDM4B through a positive feedback mechanism.Exp Cell Res. 2019 Jan 1;374(1):221-230. doi: 10.1016/j.yexcr.2018.11.027. Epub 2018 Nov 29. Exp Cell Res. 2019. PMID: 30503866
-
[The Mechanism of GREM1's Effect on Osteogenic/Odontogenic Differentiation of Stem Cells from Apical Papilla].Sichuan Da Xue Xue Bao Yi Xue Ban. 2021 May;52(3):409-415. doi: 10.12182/20210560206. Sichuan Da Xue Xue Bao Yi Xue Ban. 2021. PMID: 34018358 Free PMC article. Chinese.
-
KDM1A regulated the osteo/dentinogenic differentiation process of the stem cells of the apical papilla via binding with PLOD2.Cell Prolif. 2018 Aug;51(4):e12459. doi: 10.1111/cpr.12459. Epub 2018 Apr 15. Cell Prolif. 2018. PMID: 29656462 Free PMC article.
-
WIF1 enhanced dentinogenic differentiation in stem cells from apical papilla.BMC Oral Health. 2019 Jan 28;19(1):25. doi: 10.1186/s12903-018-0700-6. BMC Oral Health. 2019. PMID: 30691423 Free PMC article.
-
Epigallocatechin-3-Gallate Promotes Osteo-/Odontogenic Differentiation of Stem Cells from the Apical Papilla through Activating the BMP-Smad Signaling Pathway.Molecules. 2021 Mar 12;26(6):1580. doi: 10.3390/molecules26061580. Molecules. 2021. PMID: 33809391 Free PMC article.
Cited by
-
miR-140-3p enhanced the osteo/odontogenic differentiation of DPSCs via inhibiting KMT5B under hypoxia condition.Int J Oral Sci. 2021 Dec 7;13(1):41. doi: 10.1038/s41368-021-00148-y. Int J Oral Sci. 2021. PMID: 34876565 Free PMC article.
-
EZH2, via an association with KDM2B, modulates osteogenic differentiation of root apical papillary stem cells.World J Stem Cells. 2025 Apr 26;17(4):103482. doi: 10.4252/wjsc.v17.i4.103482. World J Stem Cells. 2025. PMID: 40308879 Free PMC article.
-
DNA Methylation and Histone Modification in Dental-derived Mesenchymal Stem Cells.Stem Cell Rev Rep. 2022 Dec;18(8):2797-2816. doi: 10.1007/s12015-022-10413-0. Epub 2022 Jul 27. Stem Cell Rev Rep. 2022. PMID: 35896859 Review.
-
Epigenetic control of dental stem cells: progress and prospects in multidirectional differentiation.Epigenetics Chromatin. 2024 Dec 3;17(1):37. doi: 10.1186/s13072-024-00563-5. Epigenetics Chromatin. 2024. PMID: 39623487 Free PMC article. Review.
-
Epigenetic regulation of dental-derived stem cells and their application in pulp and periodontal regeneration.PeerJ. 2023 Jan 3;11:e14550. doi: 10.7717/peerj.14550. eCollection 2023. PeerJ. 2023. PMID: 36620748 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

