Inducible TDG knockout models to study epigenetic regulation
- PMID: 33082936
- PMCID: PMC7527862
- DOI: 10.12688/f1000research.25637.2
Inducible TDG knockout models to study epigenetic regulation
Abstract
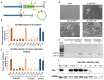
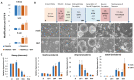
Mechanistic and functional studies by gene disruption or editing approaches often suffer from confounding effects like compensatory cellular adaptations generated by clonal selection. These issues become particularly relevant when studying factors directly involved in genetic or epigenetic maintenance. To provide a genetic tool for functional and mechanistic investigation of DNA-repair mediated active DNA demethylation, we generated experimental models in mice and murine embryonic stem cells (ESCs) based on a minigene of the thymine-DNA glycosylase (TDG). The loxP-flanked miniTdg is rapidly and reliably excised in mice and ESCs by tamoxifen-induced Cre activation, depleting TDG to undetectable levels within 24 hours. We describe the functionality of the engineered miniTdg in mouse and ESCs (TDGiKO ESCs) and validate the pluripotency and differentiation potential of TDGiKO ESCs as well as the phenotype of induced TDG depletion. The controlled and rapid depletion of TDG allows for a precise manipulation at any point in time of multistep experimental procedures as presented here for neuronal differentiation in vitro. Thus, we provide a tested and well-controlled genetic tool for the functional and mechanistic investigation of TDG in active DNA (de)methylation and/or DNA repair with minimal interference from adaptive effects and clonal selection.
Keywords: Active DNA Demethylation; Base Excision Repair; Cre/loxP; Embryonic Stem Cells; Minigene; Neuronal Differentiation; TDG; Tamoxifen.
Copyright: © 2020 Schwarz SD et al.
Conflict of interest statement
No competing interests were disclosed.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

