Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 Spike immunogen induces potent humoral and cellular immune responses
- PMID: 33083026
- PMCID: PMC7522255
- DOI: 10.1038/s41541-020-00243-x
Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 Spike immunogen induces potent humoral and cellular immune responses
Abstract
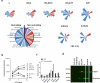
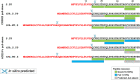
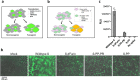
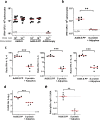
Development of effective preventative interventions against SARS-CoV-2, the etiologic agent of COVID-19 is urgently needed. The viral surface spike (S) protein of SARS-CoV-2 is a key target for prophylactic measures as it is critical for the viral replication cycle and the primary target of neutralizing antibodies. We evaluated design elements previously shown for other coronavirus S protein-based vaccines to be successful, e.g., prefusion-stabilizing substitutions and heterologous signal peptides, for selection of a S-based SARS-CoV-2 vaccine candidate. In vitro characterization demonstrated that the introduction of stabilizing substitutions (i.e., furin cleavage site mutations and two consecutive prolines in the hinge region of S2) increased the ratio of neutralizing versus non-neutralizing antibody binding, suggestive for a prefusion conformation of the S protein. Furthermore, the wild-type signal peptide was best suited for the correct cleavage needed for a natively folded protein. These observations translated into superior immunogenicity in mice where the Ad26 vector encoding for a membrane-bound stabilized S protein with a wild-type signal peptide elicited potent neutralizing humoral immunity and cellular immunity that was polarized towards Th1 IFN-γ. This optimized Ad26 vector-based vaccine for SARS-CoV-2, termed Ad26.COV2.S, is currently being evaluated in a phase I clinical trial (ClinicalTrials.gov Identifier: NCT04436276).
Keywords: SARS-CoV-2; Vaccines; Viral infection.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsThe authors declare no competing financial interests. R.B., L.R., M.J.G.B., F.W., D.Z., and J.P.L. are co-inventors on related vaccine patents. R.B., L.R., J.E.M.vd.L., M.J.G.B., G.H., F.W., D.Z., A.H.d.W., A.K., A.V., D.v.M., T.K., R.V., J.V., J.P.M.L., R.C.Z, J.C., and H.S. are employees of Janssen Vaccines & Prevention BV. R.B., L.R., F.W., D.Z., D.v.M., T.K., R.V., J.V., J.P.M.L., R.C.Z, J.C., and H.S. hold stock of Johnson & Johnson.
Figures



References
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

