Insight into an Oxidative DNA-Cleaving DNAzyme: Multiple Cofactors, the Catalytic Core Map and a Highly Efficient Variant
- PMID: 33083724
- PMCID: PMC7522124
- DOI: 10.1016/j.isci.2020.101555
Insight into an Oxidative DNA-Cleaving DNAzyme: Multiple Cofactors, the Catalytic Core Map and a Highly Efficient Variant
Abstract
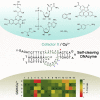
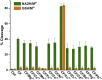
An oxidative DNA-cleaving DNAzyme (PL) employs a double-cofactor model "X/Cu2+" for catalysis. Herein, we verified that reduced nicotinamide adenine dinucleotide (NADH), flavin mononucleotide, cysteine, dithiothreitol, catechol, resorcinol, hydroquinone, phloroglucinol, o-phenylenediamine, 3,3',5,5'-tetramethylbenzidine, and hydroxylamine acted as cofactor X. According to their structural similarities or fluorescence property, we further confirmed that reduced nicotinamide adenine dinucleotide phosphate (NADPH), 2-mercaptoethanol, dopamine, chlorogenic acid, resveratrol, and 5-carboxyfluorescein also functioned as cofactor X. Superoxide anions might be the commonality behind these cofactors. We subsequently determined the conservative change of individual nucleotides in the catalytic core under four different cofactor X. The nucleotides A4 and C5 are highly conserved, whereas the conservative levels of other nucleotides are dependent on the types of cofactor X. Moreover, we observed that the minor change in the PL's secondary structure affects electrophoretic mobility. Finally, we characterized a highly efficient variant T3G and converted its double-cofactor NADH/Cu2+ to sole-cofactor NADH.
Keywords: Biocatalysis; Biochemistry; Chemistry.
© 2020 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
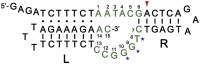




Similar articles
-
The Triple Roles of Glutathione for a DNA-Cleaving DNAzyme and Development of a Fluorescent Glutathione/Cu2+-Dependent DNAzyme Sensor for Detection of Cu2+ in Drinking Water.ACS Sens. 2017 Mar 24;2(3):364-370. doi: 10.1021/acssensors.6b00667. Epub 2017 Feb 23. ACS Sens. 2017. PMID: 28723208
-
New cofactors and inhibitors for a DNA-cleaving DNAzyme: superoxide anion and hydrogen peroxide mediated an oxidative cleavage process.Sci Rep. 2017 Mar 23;7(1):378. doi: 10.1038/s41598-017-00329-y. Sci Rep. 2017. PMID: 28336968 Free PMC article.
-
A hydrogen bond network in the active site of Anabaena ferredoxin-NADP(+) reductase modulates its catalytic efficiency.Biochim Biophys Acta. 2014 Feb;1837(2):251-63. doi: 10.1016/j.bbabio.2013.10.010. Epub 2013 Nov 4. Biochim Biophys Acta. 2014. PMID: 24200908
-
Production of riboflavin and related cofactors by biotechnological processes.Microb Cell Fact. 2020 Feb 13;19(1):31. doi: 10.1186/s12934-020-01302-7. Microb Cell Fact. 2020. PMID: 32054466 Free PMC article. Review.
-
Biomimetic cofactors and methods for their recycling.Curr Opin Chem Biol. 2019 Apr;49:59-66. doi: 10.1016/j.cbpa.2018.10.003. Epub 2018 Oct 16. Curr Opin Chem Biol. 2019. PMID: 30336443 Review.
Cited by
-
Cleaving DNA with DNA: Cooperative Tuning of Structure and Reactivity Driven by Copper Ions.Adv Sci (Weinh). 2024 Apr;11(16):e2306710. doi: 10.1002/advs.202306710. Epub 2024 Feb 28. Adv Sci (Weinh). 2024. PMID: 38419268 Free PMC article.
-
Quantity of Cu(II) ions in a copper pot by a DNAzyme-based fluorescent sensor.Food Chem (Oxf). 2025 Feb 18;10:100249. doi: 10.1016/j.fochms.2025.100249. eCollection 2025 Jun. Food Chem (Oxf). 2025. PMID: 40084088 Free PMC article.
References
-
- Ali M.M., Aguirre S.D., Lazim H., Li Y. Fluorogenic DNAzyme probes as bacterial indicators. Angew. Chem. Int. Ed. 2011;50:3751–3754. - PubMed
-
- Ali M.M., Wolfe M., Tram K., Gu J., Filipe C.D.M., Li Y., Brennan J.D. A DNAzyme-based colorimetric paper sensor for helicobacter pylori. Angew. Chem. Int. Ed. 2019;58:9907–9911. - PubMed
-
- Banno A., Higashi S., Shibata A., Ikeda M. A stimuli-responsive DNAzyme displaying boolean logic-gate responses. Chem. Commun. 2019;55:1959–1962. - PubMed
-
- Bolt H.M., Golka K. The debate on carcinogenicity of permanent hair dyes: new insights. Crit. Rev. Toxicol. 2007;37:521–536. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

