Spatial Transcriptomics Reveals Genes Associated with Dysregulated Mitochondrial Functions and Stress Signaling in Alzheimer Disease
- PMID: 33083725
- PMCID: PMC7522123
- DOI: 10.1016/j.isci.2020.101556
Spatial Transcriptomics Reveals Genes Associated with Dysregulated Mitochondrial Functions and Stress Signaling in Alzheimer Disease
Abstract
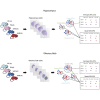
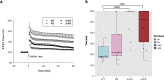
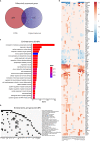
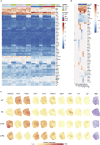
Alzheimer disease (AD) is a devastating neurological disease associated with progressive loss of mental skills and cognitive and physical functions whose etiology is not completely understood. Here, our goal was to simultaneously uncover novel and known molecular targets in the structured layers of the hippocampus and olfactory bulbs that may contribute to early hippocampal synaptic deficits and olfactory dysfunction in AD mice. Spatially resolved transcriptomics was used to identify high-confidence genes that were differentially regulated in AD mice relative to controls. A diverse set of genes that modulate stress responses and transcription were predominant in both hippocampi and olfactory bulbs. Notably, we identify Bok, implicated in mitochondrial physiology and cell death, as a spatially downregulated gene in the hippocampus of mouse and human AD brains. In summary, we provide a rich resource of spatially differentially expressed genes, which may contribute to understanding AD pathology.
Keywords: Cellular Neuroscience; Omics; Transcriptomics.
Conflict of interest statement
J.L. and J.F.N are scientific advisors at 10x Genomics Inc, which provides commercial barcoded arrays. All other authors declare no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

