Single-Chain Lanthanide Luminescence Biosensors for Cell-Based Imaging and Screening of Protein-Protein Interactions
- PMID: 33083762
- PMCID: PMC7509216
- DOI: 10.1016/j.isci.2020.101533
Single-Chain Lanthanide Luminescence Biosensors for Cell-Based Imaging and Screening of Protein-Protein Interactions
Abstract
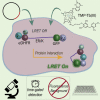
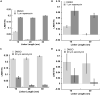
Lanthanide-based, Förster resonance energy transfer (LRET) biosensors enabled sensitive, time-gated luminescence (TGL) imaging or multiwell plate analysis of protein-protein interactions (PPIs) in living cells. We prepared stable cell lines that expressed polypeptides composed of an alpha helical linker flanked by a Tb(III) complex-binding domain, GFP, and two interacting domains at each terminus. The PPIs examined included those between FKBP12 and the rapamycin-binding domain of m-Tor (FRB) and between p53 (1-92) and HDM2 (1-128). TGL microscopy revealed dramatic differences (>500%) in donor- or acceptor-denominated, Tb(III)-to-GFP LRET ratios between open (unbound) and closed (bound) states of the biosensors. We observed much larger signal changes (>2,500%) and Z'-factors of 0.5 or more when we grew cells in 96- or 384-well plates and analyzed PPI changes using a TGL plate reader. The modular design and exceptional dynamic range of lanthanide-based LRET biosensors will facilitate versatile imaging and cell-based screening of PPIs.
Keywords: Biomolecular Engineering; Molecular Interaction; Molecular Spectroscopy Techniques; Sensor.
© 2020 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Förster resonance energy transfer biosensors for fluorescence and time-gated luminescence analysis of rac1 activity.Sci Rep. 2022 Mar 28;12(1):5291. doi: 10.1038/s41598-022-09364-w. Sci Rep. 2022. PMID: 35351946 Free PMC article.
-
Lanthanide-based resonance energy transfer biosensors for live-cell applications.Methods Enzymol. 2021;651:291-311. doi: 10.1016/bs.mie.2021.01.010. Epub 2021 Feb 23. Methods Enzymol. 2021. PMID: 33888207
-
Lanthanide-based imaging of protein-protein interactions in live cells.Inorg Chem. 2014 Feb 17;53(4):1839-53. doi: 10.1021/ic4018739. Epub 2013 Oct 21. Inorg Chem. 2014. PMID: 24144069 Free PMC article. Review.
-
An adaptable luminescence resonance energy transfer assay for measuring and screening protein-protein interactions and their inhibition.Chembiochem. 2012 Mar 5;13(4):553-8, 489. doi: 10.1002/cbic.201100710. Epub 2012 Jan 23. Chembiochem. 2012. PMID: 22271654 Free PMC article.
-
Responsive Metal Complex Probes for Time-Gated Luminescence Biosensing and Imaging.Acc Chem Res. 2020 Jul 21;53(7):1316-1329. doi: 10.1021/acs.accounts.0c00172. Epub 2020 Jun 23. Acc Chem Res. 2020. PMID: 32574043 Review.
Cited by
-
Bright and stable luminescent probes for target engagement profiling in live cells.Nat Chem Biol. 2021 Nov;17(11):1168-1177. doi: 10.1038/s41589-021-00877-5. Epub 2021 Oct 21. Nat Chem Biol. 2021. PMID: 34675420 Free PMC article.
-
High brightness red emitting polymer beads for immunoassays: Comparison between trifluoroacetylacetonates of Europium.Front Chem. 2023 Apr 20;11:1179247. doi: 10.3389/fchem.2023.1179247. eCollection 2023. Front Chem. 2023. PMID: 37153529 Free PMC article.
-
E3 Ubiquitin Ligase SPL2 Is a Lanthanide-Binding Protein.Int J Mol Sci. 2021 May 27;22(11):5712. doi: 10.3390/ijms22115712. Int J Mol Sci. 2021. PMID: 34071935 Free PMC article.
-
Förster resonance energy transfer biosensors for fluorescence and time-gated luminescence analysis of rac1 activity.Sci Rep. 2022 Mar 28;12(1):5291. doi: 10.1038/s41598-022-09364-w. Sci Rep. 2022. PMID: 35351946 Free PMC article.
References
-
- Allen M.D., Zhang J. Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. Biochem. biophys. Res. Commun. 2006;348:716–721. - PubMed
-
- Arai R., Ueda H., Kitayama A., Kamiya N., Nagamune T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. Des. Selection. 2001;14:529–532. - PubMed
-
- Aulsebrook M.L., Graham B., Grace M.R., Tuck K.L. Lanthanide complexes for luminescence-based sensing of low molecular weight analytes. Coord. Chem. Rev. 2018;375:191–220.
-
- Banaszynski L.A., Liu C.W., Wandless T.J. Characterization of the FKBP.rapamycin.FRB ternary complex. J. Am. Chem. Soc. 2005;127:4715–4721. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

