The Strategies of Pathogen-Oriented Therapy on Circumventing Antimicrobial Resistance
- PMID: 33083786
- PMCID: PMC7539235
- DOI: 10.34133/2020/2016201
The Strategies of Pathogen-Oriented Therapy on Circumventing Antimicrobial Resistance
Abstract
The emerging antimicrobial resistance (AMR) poses serious threats to the global public health. Conventional antibiotics have been eclipsed in combating with drug-resistant bacteria. Moreover, the developing and deploying of novel antimicrobial drugs have trudged, as few new antibiotics are being developed over time and even fewer of them can hit the market. Alternative therapeutic strategies to resolve the AMR crisis are urgently required. Pathogen-oriented therapy (POT) springs up as a promising approach in circumventing antibiotic resistance. The tactic underling POT is applying antibacterial compounds or materials directly to infected regions to treat specific bacteria species or strains with goals of improving the drug efficacy and reducing nontargeting and the development of drug resistance. This review exemplifies recent trends in the development of POTs for circumventing AMR, including the adoption of antibiotic-antibiotic conjugates, antimicrobial peptides, therapeutic monoclonal antibodies, nanotechnologies, CRISPR-Cas systems, and microbiota modulations. Employing these alternative approaches alone or in combination shows promising advantages for addressing the growing clinical embarrassment of antibiotics in fighting drug-resistant bacteria.
Copyright © 2020 Zifang Shang et al.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures
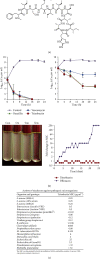
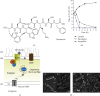
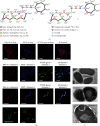

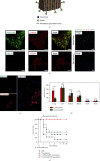

References
Publication types
LinkOut - more resources
Full Text Sources

