This is a preprint.
Progressive and accurate assembly of multi-domain protein structures from cryo-EM density maps
- PMID: 33083802
- PMCID: PMC7574260
- DOI: 10.1101/2020.10.15.340455
Progressive and accurate assembly of multi-domain protein structures from cryo-EM density maps
Update in
-
Progressive assembly of multi-domain protein structures from cryo-EM density maps.Nat Comput Sci. 2022 Apr;2(4):265-275. doi: 10.1038/s43588-022-00232-1. Epub 2022 Apr 28. Nat Comput Sci. 2022. PMID: 35844960 Free PMC article.
Abstract
Progress in cryo-electron microscopy (cryo-EM) has provided the potential for large-size protein structure determination. However, the solution rate for multi-domain proteins remains low due to the difficulty in modeling inter-domain orientations. We developed DEMO-EM, an automatic method to assemble multi-domain structures from cryo-EM maps through a progressive structural refinement procedure combining rigid-body domain fitting and flexible assembly simulations with deep neural network inter-domain distance profiles. The method was tested on a large-scale benchmark set of proteins containing up to twelve continuous and discontinuous domains with medium-to-low-resolution density maps, where DEMO-EM produced models with correct inter-domain orientations (TM-score >0.5) for 98% of cases and significantly outperformed the state-of-the-art methods. DEMO-EM was applied to SARS-Cov-2 coronavirus genome and generated models with average TM-score/RMSD of 0.97/1.4Å to the deposited structures. These results demonstrated an efficient pipeline that enables automated and reliable large-scale multi-domain protein structure modeling with atomic-level accuracy from cryo-EM maps.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Figures
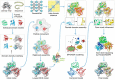
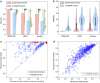

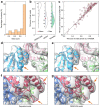

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
