Yin-Yang 1 and HBx protein activate HBV transcription by mediating the spatial interaction of cccDNA minichromosome with cellular chromosome 19p13.11
- PMID: 33084547
- PMCID: PMC7671595
- DOI: 10.1080/22221751.2020.1840311
Yin-Yang 1 and HBx protein activate HBV transcription by mediating the spatial interaction of cccDNA minichromosome with cellular chromosome 19p13.11
Abstract
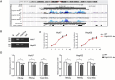
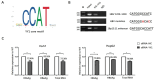
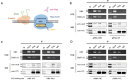
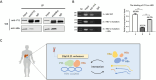
HBV cccDNA stably exists in the nuclei of infected cells as an episomal munichromosome which is responsible for viral persistence and failure of current antiviral treatments. However, the regulatory mechanism of cccDNA transcription by viral and host cellular factors is not well understood. In this study, we investigated whether cccDNA could be recruited into a specific region of the nucleus via specific interaction with a cellular chromatin to regulate its transcription activity. To investigate this hypothesis, we used chromosome conformation capture (3C) technology to search for the potential interaction of cccDNA and cellular chromatin through rcccDNA transfection in hepatoma cells and found that cccDNA is specifically associated with human chromosome 19p13.11 region, which contains a highly active enhancer element. We also confirmed that cellular transcription factor Yin-Yang 1 (YY1) and viral protein HBx mediated the spatial regulation of HBV cccDNA transcription by 19p13.11 enhancer. Thus, These findings indicate that YY1 and HBx mediate the recruitment of HBV cccDNA minichromosomes to 19p13.11 region for transcription activation, and YY1 may present as a novel therapeutic target against HBV infection.
Keywords: HBV cccDNA; HBV transcription regulation; YY1; chromosome 19p13.11; spatial interaction.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
-
- Wei XL, Luo HY, Li CF, et al. Hepatitis B virus infection is associated with younger median age at diagnosis and death in cancers. Int J Cancer. 2017;141(1):152–159. - PubMed
-
- World Health Organization . Hepatitis B 2018. Available from: http://www.who.int/news-room/fact-sheets/detail/hepatitis-b.
-
- Gane EJ. Future anti-HBV strategies. Liver Int 2017;37:40–44. - PubMed
-
- Wang J, Huang H, Liu Y, et al. HBV genome and life cycle. Adv Exp Med Biol. 2020;1179:17–37. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
