ARID1A loss in adult hepatocytes activates β-catenin-mediated erythropoietin transcription
- PMID: 33084574
- PMCID: PMC7641585
- DOI: 10.7554/eLife.53550
ARID1A loss in adult hepatocytes activates β-catenin-mediated erythropoietin transcription
Abstract
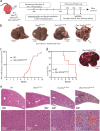
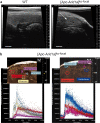
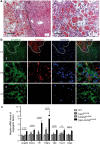
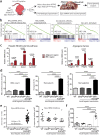
Erythropoietin (EPO) is a key regulator of erythropoiesis. The embryonic liver is the main site of erythropoietin synthesis, after which the kidney takes over. The adult liver retains the ability to express EPO, and we discovered here new players of this transcription, distinct from the classical hypoxia-inducible factor pathway. In mice, genetically invalidated in hepatocytes for the chromatin remodeler Arid1a, and for Apc, the major silencer of Wnt pathway, chromatin was more accessible and histone marks turned into active ones at the Epo downstream enhancer. Activating β-catenin signaling increased binding of Tcf4/β-catenin complex and upregulated its enhancer function. The loss of Arid1a together with β-catenin signaling, resulted in cell-autonomous EPO transcription in mouse and human hepatocytes. In mice with Apc-Arid1a gene invalidations in single hepatocytes, Epo de novo synthesis led to its secretion, to splenic erythropoiesis and to dramatic erythrocytosis. Thus, we identified new hepatic EPO regulation mechanism stimulating erythropoiesis.
Keywords: Wnt signaling; chromatin remodeling; chromosomes; developmental biology; epo transcription; gene expression; human; liver physiopathology; mouse.
© 2020, Riou et al.
Conflict of interest statement
RR, ML, SG, PB, AG, CG, RL, IL, FL, JC, AD, ZW, FV, SC No competing interests declared
Figures



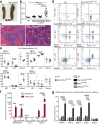

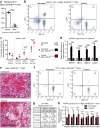


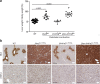
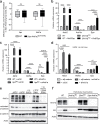




References
-
- Anson M, Crain-Denoyelle AM, Baud V, Chereau F, Gougelet A, Terris B, Yamagoe S, Colnot S, Viguier M, Perret C, Couty JP. Oncogenic β-catenin triggers an inflammatory response that determines the aggressiveness of hepatocellular carcinoma in mice. Journal of Clinical Investigation. 2012;122:586–599. doi: 10.1172/JCI43937. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- Labex Who Am I"; Idex "EpilivCan""/Agence Nationale de la Recherche/International
- Equipe Labellisée/Ligue Contre le Cancer/International
- Idex "EpilivCan"/Agence Nationale de la Recherche/International
- Chromaliv/Institut National Du Cancer/International
- Epigenetics and Liver Cancer/Institut National Du Cancer/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

