Building a Pharmacogenomics Knowledge Model Toward Precision Medicine: Case Study in Melanoma
- PMID: 33084582
- PMCID: PMC7641779
- DOI: 10.2196/20291
Building a Pharmacogenomics Knowledge Model Toward Precision Medicine: Case Study in Melanoma
Abstract
Background: Many drugs do not work the same way for everyone owing to distinctions in their genes. Pharmacogenomics (PGx) aims to understand how genetic variants influence drug efficacy and toxicity. It is often considered one of the most actionable areas of the personalized medicine paradigm. However, little prior work has included in-depth explorations and descriptions of drug usage, dosage adjustment, and so on.
Objective: We present a pharmacogenomics knowledge model to discover the hidden relationships between PGx entities such as drugs, genes, and diseases, especially details in precise medication.
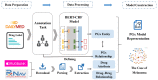
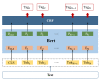
Methods: PGx open data such as DrugBank and RxNorm were integrated in this study, as well as drug labels published by the US Food and Drug Administration. We annotated 190 drug labels manually for entities and relationships. Based on the annotation results, we trained 3 different natural language processing models to complete entity recognition. Finally, the pharmacogenomics knowledge model was described in detail.
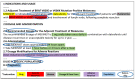
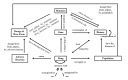
Results: In entity recognition tasks, the Bidirectional Encoder Representations from Transformers-conditional random field model achieved better performance with micro-F1 score of 85.12%. The pharmacogenomics knowledge model in our study included 5 semantic types: drug, gene, disease, precise medication (population, daily dose, dose form, frequency, etc), and adverse reaction. Meanwhile, 26 semantic relationships were defined in detail. Taking melanoma caused by a BRAF gene mutation into consideration, the pharmacogenomics knowledge model covered 7 related drugs and 4846 triples were established in this case. All the corpora, relationship definitions, and triples were made publically available.
Conclusions: We highlighted the pharmacogenomics knowledge model as a scalable framework for clinicians and clinical pharmacists to adjust drug dosage according to patient-specific genetic variation, and for pharmaceutical researchers to develop new drugs. In the future, a series of other antitumor drugs and automatic relation extractions will be taken into consideration to further enhance our framework with more PGx linked data.
Keywords: BERT–CRF model; knowledge model; melanoma; named entity recognition; pharmacogenomics.
©Hongyu Kang, Jiao Li, Meng Wu, Liu Shen, Li Hou. Originally published in JMIR Medical Informatics (http://medinform.jmir.org), 21.10.2020.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures


Similar articles
-
A Fine-Tuned Bidirectional Encoder Representations From Transformers Model for Food Named-Entity Recognition: Algorithm Development and Validation.J Med Internet Res. 2021 Aug 9;23(8):e28229. doi: 10.2196/28229. J Med Internet Res. 2021. PMID: 34383671 Free PMC article.
-
Automatic knowledge extraction from Chinese electronic medical records and rheumatoid arthritis knowledge graph construction.Quant Imaging Med Surg. 2023 Jun 1;13(6):3873-3890. doi: 10.21037/qims-22-1158. Epub 2023 May 8. Quant Imaging Med Surg. 2023. PMID: 37284084 Free PMC article.
-
Adversarial active learning for the identification of medical concepts and annotation inconsistency.J Biomed Inform. 2020 Aug;108:103481. doi: 10.1016/j.jbi.2020.103481. Epub 2020 Jul 18. J Biomed Inform. 2020. PMID: 32687985
-
Evaluation of a prototype machine learning tool to semi-automate data extraction for systematic literature reviews.Syst Rev. 2023 Oct 6;12(1):187. doi: 10.1186/s13643-023-02351-w. Syst Rev. 2023. PMID: 37803451 Free PMC article.
-
Discussion on the optimization of personalized medication using information systems based on pharmacogenomics: an example using colorectal cancer.Front Pharmacol. 2025 Jan 14;15:1516469. doi: 10.3389/fphar.2024.1516469. eCollection 2024. Front Pharmacol. 2025. PMID: 39877392 Free PMC article. Review.
References
-
- Prasad K. Role of regulatory agencies in translating pharmacogenetics to the clinics. Clin Cases Miner Bone Metab. 2009 Jan;6(1):29–34. http://europepmc.org/abstract/MED/22461095 - PMC - PubMed
-
- Table of Pharmacogenomic Biomarkers in Drug Labeling. [2020-04-27]. https://www.fda.gov/drugs/science-research-drugs/table-pharmacogenomic-b....
-
- Hida T, Nokihara H, Kondo M, Kim YH, Azuma K, Seto T, Takiguchi Y, Nishio M, Yoshioka H, Imamura F, Hotta K, Watanabe S, Goto K, Satouchi M, Kozuki T, Shukuya T, Nakagawa K, Mitsudomi T, Yamamoto N, Asakawa T, Asabe R, Tanaka T, Tamura T. Alectinib versus crizotinib in patients with ALK -positive non-small-cell lung cancer (J-ALEX): an open-label, randomised phase 3 trial. The Lancet. 2017 Jul;390(10089):29–39. doi: 10.1016/s0140-6736(17)30565-2. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

