The prevalent I686T human variant and loss-of-function mutations in the cardiomyocyte-specific kinase gene TNNI3K cause adverse contractility and concentric remodeling in mice
- PMID: 33084860
- PMCID: PMC7788294
- DOI: 10.1093/hmg/ddaa234
The prevalent I686T human variant and loss-of-function mutations in the cardiomyocyte-specific kinase gene TNNI3K cause adverse contractility and concentric remodeling in mice
Abstract
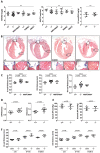
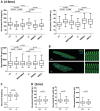
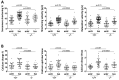
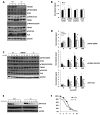
TNNI3K expression worsens disease progression in several mouse heart pathology models. TNNI3K expression also reduces the number of diploid cardiomyocytes, which may be detrimental to adult heart regeneration. However, the gene is evolutionarily conserved, suggesting a beneficial function that has remained obscure. Here, we show that C57BL/6J-inbred Tnni3k mutant mice develop concentric remodeling, characterized by ventricular wall thickening and substantial reduction of cardiomyocyte aspect ratio. This pathology occurs in mice carrying a Tnni3k null allele, a K489R point mutation rendering the protein kinase-dead, or an allele corresponding to human I686T, the most common human non-synonymous TNNI3K variant, which is hypomorphic for kinase activity. Mutant mice develop these conditions in the absence of fibrosis or hypertension, implying a primary cardiomyocyte etiology. In culture, mutant cardiomyocytes were impaired in contractility and calcium dynamics and in protein kinase A signaling in response to isoproterenol, indicating diminished contractile reserve. These results demonstrate a beneficial function of TNNI3K in the adult heart that might explain its evolutionary conservation and imply that human TNNI3K variants, in particular the widespread I686T allele, may convey elevated risk for altered heart geometry and hypertrophy.
© The Author(s) 2020. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

Similar articles
-
The Diverse Roles of TNNI3K in Cardiac Disease and Potential for Treatment.Int J Mol Sci. 2021 Jun 15;22(12):6422. doi: 10.3390/ijms22126422. Int J Mol Sci. 2021. PMID: 34203974 Free PMC article. Review.
-
Tnni3k alleles influence ventricular mononuclear diploid cardiomyocyte frequency.PLoS Genet. 2019 Oct 7;15(10):e1008354. doi: 10.1371/journal.pgen.1008354. eCollection 2019 Oct. PLoS Genet. 2019. PMID: 31589606 Free PMC article.
-
Inhibition of the cardiomyocyte-specific kinase TNNI3K limits oxidative stress, injury, and adverse remodeling in the ischemic heart.Sci Transl Med. 2013 Oct 16;5(207):207ra141. doi: 10.1126/scitranslmed.3006479. Sci Transl Med. 2013. PMID: 24132636 Free PMC article.
-
TNNI3K, a cardiac-specific kinase, promotes physiological cardiac hypertrophy in transgenic mice.PLoS One. 2013;8(3):e58570. doi: 10.1371/journal.pone.0058570. Epub 2013 Mar 5. PLoS One. 2013. PMID: 23472207 Free PMC article.
-
Troponin I-interacting protein kinase: a novel cardiac-specific kinase, emerging as a molecular target for the treatment of cardiac disease.Circ J. 2014;78(7):1514-9. doi: 10.1253/circj.cj-14-0543. Epub 2014 Jun 5. Circ J. 2014. PMID: 24899531 Free PMC article. Review.
Cited by
-
Epicardial deletion of Sox9 leads to myxomatous valve degeneration and identifies Cd109 as a novel gene associated with valve development.J Mol Cell Cardiol. 2024 Jan;186:16-30. doi: 10.1016/j.yjmcc.2023.11.002. Epub 2023 Nov 5. J Mol Cell Cardiol. 2024. PMID: 37935281 Free PMC article.
-
A kinase-dead natural polymorphism in the canine Tnni3k gene.MicroPubl Biol. 2024 May 16;2024:10.17912/micropub.biology.001164. doi: 10.17912/micropub.biology.001164. eCollection 2024. MicroPubl Biol. 2024. PMID: 38828440 Free PMC article.
-
The Diverse Roles of TNNI3K in Cardiac Disease and Potential for Treatment.Int J Mol Sci. 2021 Jun 15;22(12):6422. doi: 10.3390/ijms22126422. Int J Mol Sci. 2021. PMID: 34203974 Free PMC article. Review.
-
Loss of function of the nuclear envelope protein LEMD2 causes DNA damage-dependent cardiomyopathy.J Clin Invest. 2022 Nov 15;132(22):e158897. doi: 10.1172/JCI158897. J Clin Invest. 2022. PMID: 36377660 Free PMC article.
-
RBPMS regulates cardiomyocyte contraction and cardiac function through RNA alternative splicing.Cardiovasc Res. 2024 Feb 27;120(1):56-68. doi: 10.1093/cvr/cvad166. Cardiovasc Res. 2024. PMID: 37890031 Free PMC article.
References
-
- Zhao Y., Meng X.M., Wei Y.J., Zhao X.W., Liu D.Q., Cao H.Q., Liew C.C. and Ding J.F. (2003) Cloning and characterization of a novel cardiac-specific kinase that interacts specifically with cardiac troponin I. J. Mol. Med. (Berl), 81, 297–304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

