Viral pathogen-induced mechanisms to antagonize mammalian interferon (IFN) signaling pathway
- PMID: 33084946
- PMCID: PMC7576986
- DOI: 10.1007/s00018-020-03671-z
Viral pathogen-induced mechanisms to antagonize mammalian interferon (IFN) signaling pathway
Abstract
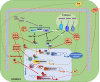
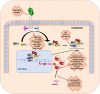
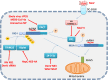
Antiviral responses of interferons (IFNs) are crucial in the host immune response, playing a relevant role in controlling viralw infections. Three types of IFNs, type I (IFN-α, IFN-β), II (IFN-γ) and III (IFN-λ), are classified according to their receptor usage, mode of induction, biological activity and amino acid sequence. Here, we provide a comprehensive review of type I IFN responses and different mechanisms that viruses employ to circumvent this response. In the first part, we will give an overview of the different induction and signaling cascades induced in the cell by IFN-I after virus encounter. Next, highlights of some of the mechanisms used by viruses to counteract the IFN induction will be described. And finally, we will address different mechanism used by viruses to interference with the IFN signaling cascade and the blockade of IFN induced antiviral activities.
Keywords: Antiviral responses; IFN-I pathway; Viral evasion mechanisms.
Figures
References
-
- Isaacs A, Lindenmann J (1957) Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci 147 (927):258–267. doi:10.1098/rspb.1957.0048 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

