The WNT/β-catenin dependent transcription: A tissue-specific business
- PMID: 33085215
- PMCID: PMC9285942
- DOI: 10.1002/wsbm.1511
The WNT/β-catenin dependent transcription: A tissue-specific business
Abstract
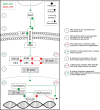
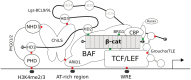
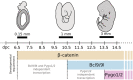
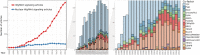
β-catenin-mediated Wnt signaling is an ancient cell-communication pathway in which β-catenin drives the expression of certain genes as a consequence of the trigger given by extracellular WNT molecules. The events occurring from signal to transcription are evolutionarily conserved, and their final output orchestrates countless processes during embryonic development and tissue homeostasis. Importantly, a dysfunctional Wnt/β-catenin pathway causes developmental malformations, and its aberrant activation is the root of several types of cancer. A rich literature describes the multitude of nuclear players that cooperate with β-catenin to generate a transcriptional program. However, a unified theory of how β-catenin drives target gene expression is still missing. We will discuss two types of β-catenin interactors: transcription factors that allow β-catenin to localize at target regions on the DNA, and transcriptional co-factors that ultimately activate gene expression. In contrast to the presumed universality of β-catenin's action, the ensemble of available evidence suggests a view in which β-catenin drives a complex system of responses in different cells and tissues. A malleable armamentarium of players might interact with β-catenin in order to activate the right "canonical" targets in each tissue, developmental stage, or disease context. Discovering the mechanism by which each tissue-specific β-catenin response is executed will be crucial to comprehend how a seemingly universal pathway fosters a wide spectrum of processes during development and homeostasis. Perhaps more importantly, this could ultimately inform us about which are the tumor-specific components that need to be targeted to dampen the activity of oncogenic β-catenin. This article is categorized under: Cancer > Molecular and Cellular Physiology Cancer > Genetics/Genomics/Epigenetics Cancer > Stem Cells and Development.
Keywords: Wnt signaling; cell signaling; genomics; transcription; β-catenin.
© 2020 The Authors. WIREs Systems Biology and Medicine published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare the absence of any conflict of interest.
Figures



References
-
- Albuquerque, C. , Breukel, C. , van der Liijt, R. , Fidalgo, P. , Lage, P. , Slors, F. J. M. , … Smits, R. (2002). The “just‐right” signaling model: APC somatic mutations are selected based on a specific level of activation of the beta‐catenin signaling cascade. Human Molecular Genetics, 11(13), 1549–1560. 10.1093/hmg/11.13.1549 - DOI - PubMed
-
- Andrews, P. G. P. , Lake, B. B. , Popadiuk, C. , & Kao, K. R. (2007). Requirement of Pygopus 2 in breast cancer. International Journal of Oncology, 30(2), 357–363 http://www.ncbi.nlm.nih.gov/pubmed/17203217 - PubMed
-
- Archbold, H. C. , Broussard, C. , Chang, M. V. , & Cadigan, K. M. (2014). Bipartite recognition of DNA by TCF/pangolin is remarkably flexible and contributes to transcriptional responsiveness and tissue specificity of wingless signaling. PLoS Genetics, 10(9), e1004591. 10.1371/journal.pgen.1004591 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

