CircRNA EPHB4 modulates stem properties and proliferation of gliomas via sponging miR-637 and up-regulating SOX10
- PMID: 33085838
- PMCID: PMC7858283
- DOI: 10.1002/1878-0261.12830
CircRNA EPHB4 modulates stem properties and proliferation of gliomas via sponging miR-637 and up-regulating SOX10
Retraction in
-
RETRACTION: CircRNA EPHB4 modulates stem properties and proliferation of gliomas via sponging miR-637 and up-regulating SOX10.Mol Oncol. 2024 Oct;18(10):2603. doi: 10.1002/1878-0261.13693. Epub 2024 Jul 1. Mol Oncol. 2024. PMID: 38952175 Free PMC article.
Abstract
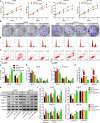
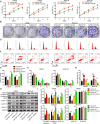
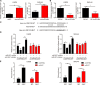
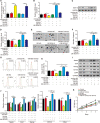
Gliomas are the most common type of primary brain tumors. CircRNA ephrin type-B receptor 4 (circEPHB4) is a circular RNA derived from the receptor tyrosine kinase EPHB4. However, the clinical significance and the specific roles of circEPHB4 in gliomas and glioma cancer stem cells (CSC) have not been studied. Here, we found that circEPHB4 (hsa_circ_0081519) and SOX10 were up-regulated and microRNA (miR)-637 was down-regulated in glioma tissues and cell lines. Consistently, circEPHB4 was positively correlated with SOX10 but negatively correlated with miR-637. The altered expressions of these molecules were independently associated with overall survival of patients. CircEPHB4 up-regulated SOX10 and Nestin by directly sponging miR-637, thereby stimulating stemness, proliferation and glycolysis of glioma cells. Functionally, silencing circEPHB4 or increasing miR-637 levels in glioma cells was sufficient to inhibit xenograft growth in vivo. In conclusion, the circEPHB4/miR-637/SOX10/Nestin axis plays a central role in controlling stem properties, self-renewal and glycolysis of glioma cells and predicts the overall survival of glioma patients. Targeting this axis might provide a therapeutic strategy for malignant gliomas.
Keywords: SOX10; cancer stemness; circEPHB4; gliomas; miR-637; proliferation.
© 2020 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA & Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68, 394–424. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

