Direct comparison of five different 3D extracellular matrix model systems for characterization of cancer cell migration
- PMID: 33085847
- PMCID: PMC7941507
- DOI: 10.1002/cnr2.1257
Direct comparison of five different 3D extracellular matrix model systems for characterization of cancer cell migration
Abstract
Background: Three-dimensional (3D) in vitro model systems can bridge the gap between regular two-dimensional cell culture and whole-animal studies. Analyses of cancer cell migration and invasion increasingly use differing 3D systems, which may produce conflicting findings.
Aims: We directly compared different 3D extracellular matrix systems for studying cancer cell migration/invasion by analyzing cell morphologies and quantifying aspects of cell migration including speed and directional persistence using automated computer-based cell tracking.
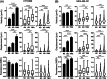
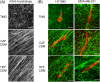
Methods and results: We performed direct comparisons of five different 3D extracellular matrix cell culture systems using both HT1080 fibrosarcoma and MDA-MB-231 breast carcinoma cell lines. The reconstituted 3D systems included two types of collagen hydrogel and tissue matrix gel (TMG) vs cell-derived matrices extracted from cultured primary human or cancer-associated fibroblasts. The fibrillar matrix architecture of these systems differed. 3D rat tail collagen and TMG matrices had short, randomly oriented collagen fibrils; bovine collagen had long, larger fibril bundles; and the cell-derived matrices were strongly oriented. HT1080 cells displayed rounded morphologies in all three reconstituted 3D matrices but became spindle shaped in the two cell-derived matrices. MDA-MB-231 cell morphologies were elongated in all matrices. Quantitative measures of cell migration parameters differed markedly between the different types of 3D matrix. Comparing the reconstituted matrices, cells migrated the most rapidly and furthest in TMG. Comparing TMG with cell-derived matrices, cells migrated more efficiently in the cell-derived matrices. The most notable differences were in directional persistence of migration, which was greatest in the two cell-derived matrices.
Conclusion: The morphologies of matrix fibrils and cell shape, and particularly the efficiency and directionality of cell migration, differed substantially depending on the type of 3D matrix system. We suggest that it is important to employ the 3D model system that most closely resembles the matrix environment being studied for analyses of cancer cell migration and invasion.
Keywords: cancer; cell migration; collagen; extracellular matrix; invasion; three-dimensional culture.
© 2020 Wiley Periodicals LLC.
Conflict of interest statement
The authors have no conflict of interest to report.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

