CMTR1-Catalyzed 2'-O-Ribose Methylation Controls Neuronal Development by Regulating Camk2α Expression Independent of RIG-I Signaling
- PMID: 33086056
- PMCID: PMC7574844
- DOI: 10.1016/j.celrep.2020.108269
CMTR1-Catalyzed 2'-O-Ribose Methylation Controls Neuronal Development by Regulating Camk2α Expression Independent of RIG-I Signaling
Abstract
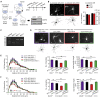
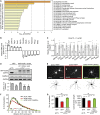
Eukaryotic mRNAs are 5' end capped with a 7-methylguanosine, which is important for processing and translation of mRNAs. Cap methyltransferase 1 (CMTR1) catalyzes 2'-O-ribose methylation of the first transcribed nucleotide (N1 2'-O-Me) to mask mRNAs from innate immune surveillance by retinoic-acid-inducible gene-I (RIG-I). Nevertheless, whether this modification regulates gene expression for neuronal functions remains unexplored. Here, we find that knockdown of CMTR1 impairs dendrite development independent of secretory cytokines and RIG-I signaling. Using transcriptomic analyses, we identify altered gene expression related to dendrite morphogenesis instead of RIG-I-activated interferon signaling, such as decreased calcium/calmodulin-dependent protein kinase 2α (Camk2α). In line with these molecular changes, dendritic complexity in CMTR1-insufficient neurons is rescued by ectopic expression of CaMK2α but not by inactivation of RIG-I signaling. We further generate brain-specific CMTR1-knockout mice to validate these findings in vivo. Our study reveals the indispensable role of CMTR1-catalyzed N1 2'-O-Me in gene regulation for brain development.
Keywords: 2′-O-ribose methylation; CMTR; CaMK2; RIG-I; cap1 modification; dendrite development; innate immunity.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures



Similar articles
-
Essential roles of RNA cap-proximal ribose methylation in mammalian embryonic development and fertility.Cell Rep. 2023 Jul 25;42(7):112786. doi: 10.1016/j.celrep.2023.112786. Epub 2023 Jul 11. Cell Rep. 2023. PMID: 37436893
-
CMTR1-catalyzed 2'-O-methylation promotes NMDA receptor signaling, long-term potentiation and memory.Prog Neurobiol. 2025 Aug;251:102802. doi: 10.1016/j.pneurobio.2025.102802. Epub 2025 Jun 25. Prog Neurobiol. 2025. PMID: 40571183
-
The mRNA Cap 2'-O-Methyltransferase CMTR1 Regulates the Expression of Certain Interferon-Stimulated Genes.mSphere. 2020 May 13;5(3):e00202-20. doi: 10.1128/mSphere.00202-20. mSphere. 2020. PMID: 32404510 Free PMC article.
-
Regulation and function of CMTR1-dependent mRNA cap methylation.Wiley Interdiscip Rev RNA. 2017 Nov;8(6):e1450. doi: 10.1002/wrna.1450. Epub 2017 Oct 2. Wiley Interdiscip Rev RNA. 2017. PMID: 28971629 Free PMC article. Review.
-
The RNA cap methyltransferases RNMT and CMTR1 co-ordinate gene expression during neural differentiation.Biochem Soc Trans. 2023 Jun 28;51(3):1131-1141. doi: 10.1042/BST20221154. Biochem Soc Trans. 2023. PMID: 37145036 Free PMC article. Review.
Cited by
-
CMTr mediated 2'-O-ribose methylation status of cap-adjacent nucleotides across animals.RNA. 2022 Oct;28(10):1377-1390. doi: 10.1261/rna.079317.122. Epub 2022 Aug 15. RNA. 2022. PMID: 35970556 Free PMC article.
-
CMTR1 is recruited to transcription start sites and promotes ribosomal protein and histone gene expression in embryonic stem cells.Nucleic Acids Res. 2022 Mar 21;50(5):2905-2922. doi: 10.1093/nar/gkac122. Nucleic Acids Res. 2022. PMID: 35212377 Free PMC article.
-
Multi-omics analysis reveals CMTR1 upregulation in cancer and roles in ribosomal protein gene expression and tumor growth.Cell Commun Signal. 2025 Apr 24;23(1):197. doi: 10.1186/s12964-025-02147-6. Cell Commun Signal. 2025. PMID: 40275371 Free PMC article.
-
CK2 phosphorylation of CMTR1 promotes RNA cap formation and influenza virus infection.Cell Rep. 2024 Jul 23;43(7):114405. doi: 10.1016/j.celrep.2024.114405. Epub 2024 Jun 25. Cell Rep. 2024. PMID: 38923463 Free PMC article.
-
RNA Epigenetics: Fine-Tuning Chromatin Plasticity and Transcriptional Regulation, and the Implications in Human Diseases.Genes (Basel). 2021 Apr 22;12(5):627. doi: 10.3390/genes12050627. Genes (Basel). 2021. PMID: 33922187 Free PMC article. Review.
References
-
- Akichika S., Hirano S., Shichino Y., Suzuki T., Nishimasu H., Ishitani R., Sugita A., Hirose Y., Iwasaki S., Nureki O., Suzuki T. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science. 2019;363:eaav0080. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

