Assessing the Direct Binding of Ark-Like E3 RING Ligases to Ubiquitin and Its Implication on Their Protein Interaction Network
- PMID: 33086510
- PMCID: PMC7594095
- DOI: 10.3390/molecules25204787
Assessing the Direct Binding of Ark-Like E3 RING Ligases to Ubiquitin and Its Implication on Their Protein Interaction Network
Abstract
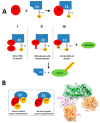
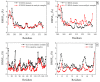
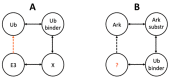
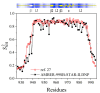
The ubiquitin pathway required for most proteins' targeted degradation involves three classes of enzymes: E1-activating enzyme, E2-conjugating enzyme, and E3-ligases. The human Ark2C is the single known E3 ligase that adopts an alternative, Ub-dependent mechanism for the activation of Ub transfer in the pathway. Its RING domain binds both E2-Ub and free Ub with high affinity, resulting in a catalytic active UbR-RING-E2-UbD complex formation. We examined potential changes in the conformational plasticity of the Ark2C RING domain and its ligands in their complexed form within the ubiquitin pathway through molecular dynamics (MD). Three molecular mechanics force fields compared to previous NMR relaxation studies of RING domain of Arkadia were used for effective and accurate assessment of MDs. Our results suggest the Ark2C Ub-RING docking site has a substantial impact on maintaining the conformational rigidity of E2-E3 assembly, necessary for the E3's catalytic activity. In the UbR-RING-E2-UbD catalytic complex, the UbR molecule was found to have greater mobility than the other Ub, bound to E2. Furthermore, network-based bioinformatics helped us identify E3 RING ligase candidates which potentially exhibit similar structural modules as Ark2C, along with predicted substrates targeted by the Ub-binding RING Ark2C. Our findings could trigger a further exploration of related unrevealed functions of various other E3 RING ligases.
Keywords: E3 RING ligases; PPI network; molecular dynamics; ubiquitin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity.Nat Struct Mol Biol. 2016 Jan;23(1):45-52. doi: 10.1038/nsmb.3142. Epub 2015 Dec 14. Nat Struct Mol Biol. 2016. PMID: 26656854
-
Exploring the RING-catalyzed ubiquitin transfer mechanism by MD and QM/MM calculations.PLoS One. 2014 Jul 8;9(7):e101663. doi: 10.1371/journal.pone.0101663. eCollection 2014. PLoS One. 2014. PMID: 25003393 Free PMC article.
-
Arkadia and Ark2C Promote Substrate Ubiquitylation with Multiple E2 Enzymes.J Mol Biol. 2025 Sep 1;437(17):169259. doi: 10.1016/j.jmb.2025.169259. Epub 2025 May 30. J Mol Biol. 2025. PMID: 40451499
-
Use of E2~ubiquitin conjugates for the characterization of ubiquitin transfer by RING E3 ligases such as the inhibitor of apoptosis proteins.Methods Enzymol. 2014;545:243-63. doi: 10.1016/B978-0-12-801430-1.00010-X. Methods Enzymol. 2014. PMID: 25065893 Review.
-
RING-Between-RING E3 Ligases: Emerging Themes amid the Variations.J Mol Biol. 2017 Nov 10;429(22):3363-3375. doi: 10.1016/j.jmb.2017.08.008. Epub 2017 Aug 19. J Mol Biol. 2017. PMID: 28827147 Free PMC article. Review.
Cited by
-
A SARS-CoV-2 -human metalloproteome interaction map.J Inorg Biochem. 2021 Jun;219:111423. doi: 10.1016/j.jinorgbio.2021.111423. Epub 2021 Mar 13. J Inorg Biochem. 2021. PMID: 33813307 Free PMC article.
References
-
- Li W., Bengtson M.H., Ulbrich A., Matsuda A., Reddy V.A., Orth A., Chanda S.K., Batalov S., Joazeiro C.A.P. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

