DNA Methylation at Birth Predicts Intellectual Functioning and Autism Features in Children with Fragile X Syndrome
- PMID: 33086711
- PMCID: PMC7589848
- DOI: 10.3390/ijms21207735
DNA Methylation at Birth Predicts Intellectual Functioning and Autism Features in Children with Fragile X Syndrome
Abstract
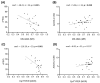
Fragile X syndrome (FXS) is a leading single-gene cause of intellectual disability (ID) with autism features. This study analysed diagnostic and prognostic utility of the Fragile X-Related Epigenetic Element 2 DNA methylation (FREE2m) assessed by Methylation Specific-Quantitative Melt Analysis and the EpiTYPER system, in retrospectively retrieved newborn blood spots (NBS) and newly created dried blood spots (DBS) from 65 children with FXS (~2-17 years). A further 168 NBS from infants from the general population were used to establish control reference ranges, in both sexes. FREE2m analysis showed sensitivity and specificity approaching 100%. In FXS males, NBS FREE2m strongly correlated with intellectual functioning and autism features, however associations were not as strong for FXS females. Fragile X mental retardation 1 gene (FMR1) mRNA levels in blood were correlated with FREE2m in both NBS and DBS, for both sexes. In females, DNAm was significantly increased at birth with a decrease in childhood. The findings support the use of FREE2m analysis in newborns for screening, diagnostic and prognostic testing in FXS.
Keywords: DNA methylation (DNAm); autism spectrum disorder (ASD); fragile X mental retardation 1 gene (FMR1 gene); fragile X syndrome (FXS); intellectual disability (ID); newborn screening.
Conflict of interest statement
David E. Godler is an inventor of the following patents: PCT/AU2010/001134; filing No. AU2010/903595; filing No. AU2011/902500; and filing No. 2013/900227, related to the technology described in this publication. David Eugeny Godler is the director of E.D.G. Innovations and Consulting Pty Ltd., Melbourne, Australia, that owns this intellectual property. The other authors declare no competing interests.
Figures


References
-
- Joubert B.R., Felix J.F., Yousefi P., Bakulski K.M., Just A.C., Breton C., Reese S.E., Markunas C.A., Richmond R.C., Xu C.-J., et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. Am. J. Hum. Genet. 2016;98:680–696. doi: 10.1016/j.ajhg.2016.02.019. - DOI - PMC - PubMed
-
- Aref-Eshghi E., Rodenhiser D.I., Schenkel L.C., Lin H., Skinner C., Ainsworth P., Paré G., Hood R.L., Bulman D.E., Kernohan K.D., et al. Genomic DNA Methylation Signatures Enable Concurrent Diagnosis and Clinical Genetic Variant Classification in Neurodevelopmental Syndromes. Am. J. Hum. Genet. 2018;102:156–174. doi: 10.1016/j.ajhg.2017.12.008. - DOI - PMC - PubMed
-
- Andrews S.V., Ellis S.E., Bakulski K.M., Sheppard B., Croen L.A., Hertz-Picciotto I., Newschaffer C.J., Feinberg A.P., Arking D.E., Ladd-Acosta C., et al. Cross-tissue integration of genetic and epigenetic data offers insight into autism spectrum disorder. Nat. Commun. 2017;8:1011. doi: 10.1038/s41467-017-00868-y. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

