History of DNA polymerase β X-ray crystallography
- PMID: 33087265
- PMCID: PMC7586737
- DOI: 10.1016/j.dnarep.2020.102928
History of DNA polymerase β X-ray crystallography
Abstract
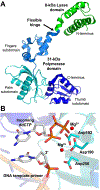
DNA polymerase β (Pol β) is an essential mammalian enzyme involved in the repair of DNA damage during the base excision repair (BER) pathway. In hopes of faithfully restoring the coding potential to damaged DNA during BER, Pol β first uses a lyase activity to remove the 5'-deoxyribose phosphate moiety from a nicked BER intermediate, followed by a DNA synthesis activity to insert a nucleotide triphosphate into the resultant 1-nucleotide gapped DNA substrate. This DNA synthesis activity of Pol β has served as a model to characterize the molecular steps of the nucleotidyl transferase mechanism used by mammalian DNA polymerases during DNA synthesis. This is in part because Pol β has been extremely amenable to X-ray crystallography, with the first crystal structure of apoenzyme rat Pol β published in 1994 by Dr. Samuel Wilson and colleagues. Since this first structure, the Wilson lab and colleagues have published an astounding 267 structures of Pol β that represent different liganded states, conformations, variants, and reaction intermediates. While many labs have made significant contributions to our understanding of Pol β, the focus of this article is on the long history of the contributions from the Wilson lab. We have chosen to highlight select seminal Pol β structures with emphasis on the overarching contributions each structure has made to the field.
Copyright © 2020 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Molecular and Functional Characteristics of DNA Polymerase Beta-Like Enzymes From Trypanosomatids.Front Cell Infect Microbiol. 2021 Aug 5;11:670564. doi: 10.3389/fcimb.2021.670564. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34422676 Free PMC article. Review.
-
DNA polymerase X of African swine fever virus: insertion fidelity on gapped DNA substrates and AP lyase activity support a role in base excision repair of viral DNA.J Mol Biol. 2003 Mar 7;326(5):1403-12. doi: 10.1016/s0022-2836(03)00019-6. J Mol Biol. 2003. PMID: 12595253
-
Comparison of functional properties of mammalian DNA polymerase lambda and DNA polymerase beta in reactions of DNA synthesis related to DNA repair.Biochim Biophys Acta. 2005 Aug 10;1751(2):150-8. doi: 10.1016/j.bbapap.2005.05.012. Biochim Biophys Acta. 2005. PMID: 15979954
-
Structure and mechanism of DNA polymerase β.Biochemistry. 2014 May 6;53(17):2768-80. doi: 10.1021/bi500139h. Epub 2014 Apr 23. Biochemistry. 2014. PMID: 24717170 Free PMC article.
-
Structural design of a eukaryotic DNA repair polymerase: DNA polymerase beta.Mutat Res. 2000 Aug 30;460(3-4):231-44. doi: 10.1016/s0921-8777(00)00029-x. Mutat Res. 2000. PMID: 10946231 Review.
Cited by
-
Molecular and Functional Characteristics of DNA Polymerase Beta-Like Enzymes From Trypanosomatids.Front Cell Infect Microbiol. 2021 Aug 5;11:670564. doi: 10.3389/fcimb.2021.670564. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34422676 Free PMC article. Review.
-
The origin of genetic and metabolic systems: Evolutionary structuralinsights.Heliyon. 2023 Mar 11;9(3):e14466. doi: 10.1016/j.heliyon.2023.e14466. eCollection 2023 Mar. Heliyon. 2023. PMID: 36967965 Free PMC article.
-
Polβ/XRCC1 heterodimerization dictates DNA damage recognition and basal Polβ protein levels without interfering with mouse viability or fertility.DNA Repair (Amst). 2023 Mar;123:103452. doi: 10.1016/j.dnarep.2023.103452. Epub 2023 Jan 20. DNA Repair (Amst). 2023. PMID: 36702010 Free PMC article.
-
Unveiling the Mechanism of Deprotonation and Proton Transfer of DNA Polymerase Catalysis via Single-Molecule Conductance.Adv Sci (Weinh). 2025 Jan;12(2):e2408112. doi: 10.1002/advs.202408112. Epub 2024 Nov 21. Adv Sci (Weinh). 2025. PMID: 39570753 Free PMC article.
-
DNA polymerase β: Closing the gap between structure and function.DNA Repair (Amst). 2020 Sep;93:102910. doi: 10.1016/j.dnarep.2020.102910. DNA Repair (Amst). 2020. PMID: 33087276 Free PMC article. Review.
References
-
- Abbotts J, SenGupta DN, Zmudzka B, Widen SG, Notario V, Wilson SH, Expression of human DNA polymerase beta in Escherichia coli and characterization of the recombinant enzyme, Biochemistry, 27 (1988) 901–909. - PubMed
-
- Sawaya MR, Pelletier H, Kumar A, Wilson SH, Kraut J, Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism, Science, 264 (1994) 1930–1935. - PubMed
-
- Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J, Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity, Biochemistry, 35 (1996) 12742–12761. - PubMed
-
- Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J, Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP, Science, 264 (1994) 1891–1903. - PubMed
Publication types
MeSH terms
Substances
Personal name as subject
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

