Human DNA ligases in replication and repair
- PMID: 33087274
- PMCID: PMC8727047
- DOI: 10.1016/j.dnarep.2020.102908
Human DNA ligases in replication and repair
Abstract
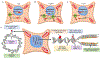
To ensure genome integrity, the joining of breaks in the phosphodiester backbone of duplex DNA is required during DNA replication and to complete the repair of almost all types of DNA damage. In human cells, this task is accomplished by DNA ligases encoded by three genes, LIG1, LIG3 and LIG4. Mutations in LIG1 and LIG4 have been identified as the causative factor in two inherited immunodeficiency syndromes. Moreover, there is emerging evidence that DNA ligases may be good targets for the development of novel anti-cancer agents. In this graphical review, we provide an overview of the roles of the DNA ligases encoded by the three human LIG genes in DNA replication and repair.
Keywords: DNA joining; Okazaki fragments; alternative end joining; base excision repair; non-homologous end joining; nucleotide excision repair; single strand break reapir.
Copyright © 2020 Elsevier B.V. All rights reserved.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

