Natural polymorphism of Ym1 regulates pneumonitis through alternative activation of macrophages
- PMID: 33087360
- PMCID: PMC7577715
- DOI: 10.1126/sciadv.aba9337
Natural polymorphism of Ym1 regulates pneumonitis through alternative activation of macrophages
Abstract
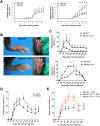
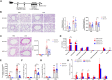
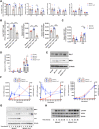
We have positionally cloned the Ym1 gene, with a duplication and a promoter polymorphism, as a major regulator of inflammation. Mice with the RIIIS/J haplotype, with the absence of Ym1 expression, showed reduced susceptibility to mannan-enhanced collagen antibody-induced arthritis and to chronic arthritis induced by intranasal exposure of mannan. Depletion of lung macrophages alleviated arthritis, whereas intranasal supplement of Ym1 protein to Ym1-deficient mice reversed the disease, suggesting a key role of Ym1 for inflammatory activity by lung macrophages. Ym1-deficient mice with pneumonitis had less eosinophil infiltration, reduced production of type II cytokines and IgG1, and skewing of macrophages toward alternative activation due to enhanced STAT6 activation. Proteomics analysis connected Ym1 polymorphism with changed lipid metabolism. Induced PPAR-γ and lipid metabolism in Ym1-deficient macrophages contributed to cellular polarization. In conclusion, the natural polymorphism of Ym1 regulates alternative activation of macrophages associated with pulmonary inflammation.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures



References
-
- Martinez F. D., Genes, environments, development and asthma: A reappraisal. Eur. Respir. J. 29, 179–184 (2007). - PubMed
-
- Olofsson P., Holmberg J., Tordsson J., Lu S., Akerstrom B., Holmdahl R., Positional identification of Ncf1 as a gene that regulates arthritis severity in rats. Nat. Genet. 33, 25–32 (2003). - PubMed
-
- Olsson L. M., Johansson A. C., Gullstrand B., Jonsen A., Saevarsdottir S., Ronnblom L., Leonard D., Wettero J., Sjowall C., Svenungsson E., Gunnarsson I., Bengtsson A. A., Holmdahl R., A single nucleotide polymorphism in the NCF1 gene leading to reduced oxidative burst is associated with systemic lupus erythematosus. Ann. Rheum. Dis. 76, 1607–1613 (2017). - PubMed
-
- Zhao J., Ma J., Deng Y., Kelly J. A., Kim K., Bang S. Y., Lee H. S., Li Q. Z., Wakeland E. K., Qiu R., Liu M., Guo J., Li Z., Tan W., Rasmussen A., Lessard C. J., Sivils K. L., Hahn B. H., Grossman J. M., Kamen D. L., Gilkeson G. S., Bae S. C., Gaffney P. M., Shen N., Tsao B. P., A missense variant in NCF1 is associated with susceptibility to multiple autoimmune diseases. Nat. Genet. 49, 433–437 (2017). - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

