The FAM171A2 gene is a key regulator of progranulin expression and modifies the risk of multiple neurodegenerative diseases
- PMID: 33087363
- PMCID: PMC7577723
- DOI: 10.1126/sciadv.abb3063
The FAM171A2 gene is a key regulator of progranulin expression and modifies the risk of multiple neurodegenerative diseases
Abstract
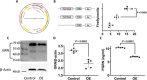
Progranulin (PGRN) is a secreted pleiotropic glycoprotein associated with the development of common neurodegenerative diseases. Understanding the pathophysiological role of PGRN may help uncover biological underpinnings. We performed a genome-wide association study to determine the genetic regulators of cerebrospinal fluid (CSF) PGRN levels. Common variants in region of FAM171A2 were associated with lower CSF PGRN levels (rs708384, P = 3.95 × 10-12). This was replicated in another independent cohort. The rs708384 was associated with increased risk of Alzheimer's disease, Parkinson's disease, and frontotemporal dementia and could modify the expression of the FAM171A2 gene. FAM171A2 was considerably expressed in the vascular endothelium and microglia, which are rich in PGRN. The in vitro study further confirmed that the rs708384 mutation up-regulated the expression of FAM171A2, which caused a decrease in the PGRN level. Collectively, genetic, molecular, and bioinformatic findings suggested that FAM171A2 is a key player in regulating PGRN production.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

