Cytidine Monophosphate N-Acetylneuraminic Acid Synthetase and Solute Carrier Family 35 Member A1 Are Required for Reovirus Binding and Infection
- PMID: 33087464
- PMCID: PMC7944449
- DOI: 10.1128/JVI.01571-20
Cytidine Monophosphate N-Acetylneuraminic Acid Synthetase and Solute Carrier Family 35 Member A1 Are Required for Reovirus Binding and Infection
Abstract
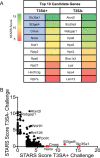
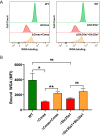
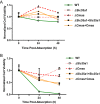
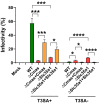
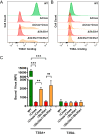
Engagement of cell surface receptors by viruses is a critical determinant of viral tropism and disease. The reovirus attachment protein σ1 binds sialylated glycans and proteinaceous receptors to mediate infection, but the specific requirements for different cell types are not entirely known. To identify host factors required for reovirus-induced cell death, we conducted a CRISPR-knockout screen targeting over 20,000 genes in murine microglial BV2 cells. Candidate genes required for reovirus to cause cell death were highly enriched for sialic acid synthesis and transport. Two of the top candidates identified, CMP N-acetylneuraminic acid synthetase (Cmas) and solute carrier family 35 member A1 (Slc35a1), promote sialic acid expression on the cell surface. Two reovirus strains that differ in the capacity to bind sialic acid, T3SA+ and T3SA-, were used to evaluate Cmas and Slc35a1 as potential host genes required for reovirus infection. Following CRISPR-Cas9 disruption of either gene, cell surface expression of sialic acid was diminished. These results correlated with decreased binding of strain T3SA+, which is capable of engaging sialic acid. Disruption of either gene did not alter the low-level binding of T3SA-, which does not engage sialic acid. Furthermore, infectivity of T3SA+ was diminished to levels similar to those of T3SA- in cells lacking Cmas and Slc35a1 by CRISPR ablation. However, exogenous expression of Cmas and Slc35a1 into the respective null cells restored sialic acid expression and T3SA+ binding and infectivity. These results demonstrate that Cmas and Slc35a1, which mediate cell surface expression of sialic acid, are required in murine microglial cells for efficient reovirus binding and infection.IMPORTANCE Attachment factors and receptors are important determinants of dissemination and tropism during reovirus-induced disease. In a CRISPR cell survival screen, we discovered two genes, Cmas and Slc35a1, which encode proteins required for sialic acid expression on the cell surface and mediate reovirus infection of microglial cells. This work elucidates host genes that render microglial cells susceptible to reovirus infection and expands current understanding of the receptors on microglial cells that are engaged by reovirus. Such knowledge may lead to new strategies to selectively target microglial cells for oncolytic applications.
Keywords: cytidine monophosphate N-acetylneuraminic acid synthetase; microglia; reovirus; sialic acid; solute carrier family 35 member A1; viral attachment.
Copyright © 2020 American Society for Microbiology.
Figures
References
-
- Tyler KL. 2001. Mammalian reoviruses, p 1729–1745. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

