Direct reprogramming of human umbilical vein- and peripheral blood-derived endothelial cells into hepatic progenitor cells
- PMID: 33087715
- PMCID: PMC7578104
- DOI: 10.1038/s41467-020-19041-z
Direct reprogramming of human umbilical vein- and peripheral blood-derived endothelial cells into hepatic progenitor cells
Abstract
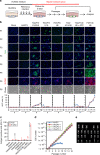
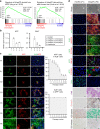
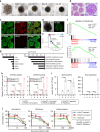
Recent advances have enabled the direct induction of human tissue-specific stem and progenitor cells from differentiated somatic cells. However, it is not known whether human hepatic progenitor cells (hHepPCs) can be generated from other cell types by direct lineage reprogramming with defined transcription factors. Here, we show that a set of three transcription factors, FOXA3, HNF1A, and HNF6, can induce human umbilical vein endothelial cells to directly acquire the properties of hHepPCs. These induced hHepPCs (hiHepPCs) propagate in long-term monolayer culture and differentiate into functional hepatocytes and cholangiocytes by forming cell aggregates and cystic epithelial spheroids, respectively, under three-dimensional culture conditions. After transplantation, hiHepPC-derived hepatocytes and cholangiocytes reconstitute damaged liver tissues and support hepatic function. The defined transcription factors also induce hiHepPCs from endothelial cells circulating in adult human peripheral blood. These expandable and bipotential hiHepPCs may be useful in the study and treatment of human liver diseases.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors.Nature. 2011 May 11;475(7356):386-9. doi: 10.1038/nature10116. Nature. 2011. PMID: 21562492
-
Chemical cocktails enable hepatic reprogramming of human urine-derived cells with a single transcription factor.Acta Pharmacol Sin. 2019 May;40(5):620-629. doi: 10.1038/s41401-018-0170-z. Epub 2018 Oct 12. Acta Pharmacol Sin. 2019. PMID: 30315254 Free PMC article.
-
Feeder-free and serum-free production of hepatocytes, cholangiocytes, and their proliferating progenitors from human pluripotent stem cells: application to liver-specific functional and cytotoxic assays.Cell Reprogram. 2012 Apr;14(2):171-85. doi: 10.1089/cell.2011.0064. Epub 2012 Mar 2. Cell Reprogram. 2012. PMID: 22384928
-
[Identification of master regulators for induction of hepatocyte differentiation].Seikagaku. 2012 Aug;84(8):675-9. Seikagaku. 2012. PMID: 23012878 Review. Japanese. No abstract available.
-
Characterization of the differentiation capacity of rat-derived hepatic stem cells.Semin Liver Dis. 2003 Nov;23(4):325-36. doi: 10.1055/s-2004-815561. Semin Liver Dis. 2003. PMID: 14722810 Review.
Cited by
-
Advances in liver organoids: replicating hepatic complexity for toxicity assessment and disease modeling.Stem Cell Res Ther. 2025 Jan 26;16(1):27. doi: 10.1186/s13287-025-04139-2. Stem Cell Res Ther. 2025. PMID: 39865320 Free PMC article. Review.
-
Generation of functional hepatocytes by forward programming with nuclear receptors.Elife. 2022 Aug 12;11:e71591. doi: 10.7554/eLife.71591. Elife. 2022. PMID: 35959725 Free PMC article.
-
The role of pioneer transcription factors in the induction of direct cellular reprogramming.Regen Ther. 2023 Jun 19;24:112-116. doi: 10.1016/j.reth.2023.06.002. eCollection 2023 Dec. Regen Ther. 2023. PMID: 37397229 Free PMC article. Review.
-
Monitoring Cultured Rat Hepatocytes Using RNA-Seq In Vitro.Int J Mol Sci. 2023 Apr 19;24(8):7534. doi: 10.3390/ijms24087534. Int J Mol Sci. 2023. PMID: 37108701 Free PMC article.
-
Transcriptional Control: A Directional Sign at the Crossroads of Adult Hepatic Progenitor Cells' Fates.Int J Biol Sci. 2024 Jun 24;20(9):3544-3556. doi: 10.7150/ijbs.93739. eCollection 2024. Int J Biol Sci. 2024. PMID: 38993564 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

