The structural basis of herpesvirus entry
- PMID: 33087881
- PMCID: PMC8579738
- DOI: 10.1038/s41579-020-00448-w
The structural basis of herpesvirus entry
Abstract
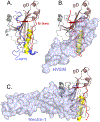
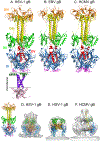
Herpesviruses are ubiquitous, double-stranded DNA, enveloped viruses that establish lifelong infections and cause a range of diseases. Entry into host cells requires binding of the virus to specific receptors, followed by the coordinated action of multiple viral entry glycoproteins to trigger membrane fusion. Although the core fusion machinery is conserved for all herpesviruses, each species uses distinct receptors and receptor-binding glycoproteins. Structural studies of the prototypical herpesviruses herpes simplex virus 1 (HSV-1), HSV-2, human cytomegalovirus (HCMV) and Epstein-Barr virus (EBV) entry glycoproteins have defined the interaction sites for glycoprotein complexes and receptors, and have revealed conformational changes that occur on receptor binding. Recent crystallography and electron microscopy studies have refined our model of herpesvirus entry into cells, clarifying both the conserved features and the unique features. In this Review, we discuss recent insights into herpesvirus entry by analysing the structures of entry glycoproteins, including the diverse receptor-binding glycoproteins (HSV-1 glycoprotein D (gD), EBV glycoprotein 42 (gp42) and HCMV gH-gL-gO trimer and gH-gL-UL128-UL130-UL131A pentamer), as well gH-gL and the fusion protein gB, which are conserved in all herpesviruses.
Figures

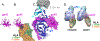

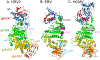
References
-
- Vallbracht M, Backovic M, Klupp BG, Rey FA & Mettenleiter TC Common characteristics and unique features: A comparison of the fusion machinery of the alphaherpesviruses Pseudorabies virus and Herpes simplex virus. Advances in virus research 104, 225–281, doi:10.1016/bs.aivir.2019.05.007 (2019). - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

