The complete chloroplast genome of the rare species Epimedium tianmenshanensis and comparative analysis with related species
- PMID: 33088051
- PMCID: PMC7548308
- DOI: 10.1007/s12298-020-00882-3
The complete chloroplast genome of the rare species Epimedium tianmenshanensis and comparative analysis with related species
Abstract
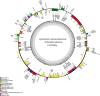
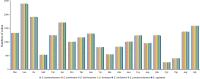
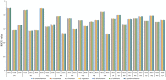
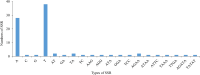
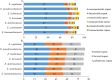
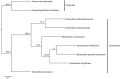
Epimedium tianmenshanensis is a rare perennial herb distributed in China, and it is also an important medicinal plant. Here, we used illumina paired-end sequencing technology to obtain the complete chloroplast genome of E. tianmenshanensis, and compared analysis with related species. The length of the complete chloroplast genome of E. tianmenshanensis is 156,956 bp, which is a relatively conserved quadripartite structure including a large single copy (LSC) region of 88,409 bp, a small single copy (SSC) region of 17,448 bp, and a pair of inverted repeat (IRa/IRb) regions of 25,550 bp. The whole genome contains 132 unique genes, including 85 protein-coding genes, 38 tRNA genes, eight rRNA genes and one pseudogene. 87 simple sequence repeats (SSRs) were identified, and most of them were found to be composed of A/T. In addition, 22,923 codons were detected in 78 protein-coding genes of E. tianmenshanensis, and the overall codon bias pattern in the genome tended to use A/U ending codons. Phylogenetic analysis demonstrated that all the Epimedium species formed a monophyletic clade, and E. tianmenshanensis had the closest relationship to E. dolichostemon. The results of this study provided useful molecular information about the evolution and molecular biology of E. tianmenshanensis.
Keywords: Chloroplast genome; Epimedium; Phylogenetic analysis; RSCU; SSRs.
© Prof. H.S. Srivastava Foundation for Science and Society 2020.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures
Similar articles
-
The complete chloroplast genome of Epimedium elachyphyllum Stearn (Berberidaceae), an endangered species endemic to China.Mitochondrial DNA B Resour. 2020 Feb 3;5(1):1027-1028. doi: 10.1080/23802359.2020.1721357. Mitochondrial DNA B Resour. 2020. PMID: 33366859 Free PMC article.
-
The complete chloroplast genome of Epimedium wushanense T. S. Ying. (Berberidaceae), a traditional Chinese medicinal herb.Mitochondrial DNA B Resour. 2020 Jan 24;5(1):817-818. doi: 10.1080/23802359.2020.1715877. Mitochondrial DNA B Resour. 2020. PMID: 33366765 Free PMC article.
-
The complete chloroplast genome of Epimedium mikinorii Stearn. (Berberidaceae).Mitochondrial DNA B Resour. 2020 Jan 24;5(1):806-807. doi: 10.1080/23802359.2020.1715863. Mitochondrial DNA B Resour. 2020. PMID: 33366760 Free PMC article.
-
The complete chloroplast genome of Epimedium davidii Franch. (Berberidaceae).Mitochondrial DNA B Resour. 2020 Jan 8;5(1):445-446. doi: 10.1080/23802359.2019.1704641. Mitochondrial DNA B Resour. 2020. PMID: 33366594 Free PMC article.
-
The complete chloroplast genome of Epimedium muhuangense (Berberidaceae).Mitochondrial DNA B Resour. 2022 May 16;7(5):870-872. doi: 10.1080/23802359.2022.2071650. eCollection 2022. Mitochondrial DNA B Resour. 2022. PMID: 35602330 Free PMC article.
Cited by
-
Complete chloroplast genome sequences of the ornamental plant Prunus cistena and comparative and phylogenetic analyses with its closely related species.BMC Genomics. 2023 Dec 5;24(1):739. doi: 10.1186/s12864-023-09838-9. BMC Genomics. 2023. PMID: 38053028 Free PMC article.
-
Three complete chloroplast genomes from two north American Rhus species and phylogenomics of Anacardiaceae.BMC Genom Data. 2024 Mar 15;25(1):30. doi: 10.1186/s12863-024-01200-6. BMC Genom Data. 2024. PMID: 38491489 Free PMC article.
-
Comparative genomics and phylogenetic analysis of six Malvaceae species based on chloroplast genomes.BMC Plant Biol. 2024 Dec 26;24(1):1245. doi: 10.1186/s12870-024-05974-w. BMC Plant Biol. 2024. PMID: 39722018 Free PMC article.
-
Comparative analysis of chloroplast genome and new insights into phylogenetic relationships of Ajuga and common adulterants.Front Plant Sci. 2023 Oct 25;14:1251829. doi: 10.3389/fpls.2023.1251829. eCollection 2023. Front Plant Sci. 2023. PMID: 37954994 Free PMC article.
-
The Complete Chloroplast Genomes of Primula obconica Provide Insight That Neither Species nor Natural Section Represent Monophyletic Taxa in Primula (Primulaceae).Genes (Basel). 2022 Mar 23;13(4):567. doi: 10.3390/genes13040567. Genes (Basel). 2022. PMID: 35456373 Free PMC article.
References
LinkOut - more resources
Full Text Sources
