Positive selection within the genomes of SARS-CoV-2 and other Coronaviruses independent of impact on protein function
- PMID: 33088633
- PMCID: PMC7571416
- DOI: 10.7717/peerj.10234
Positive selection within the genomes of SARS-CoV-2 and other Coronaviruses independent of impact on protein function
Abstract
Background: The emergence of a novel coronavirus (SARS-CoV-2) associated with severe acute respiratory disease (COVID-19) has prompted efforts to understand the genetic basis for its unique characteristics and its jump from non-primate hosts to humans. Tests for positive selection can identify apparently nonrandom patterns of mutation accumulation within genomes, highlighting regions where molecular function may have changed during the origin of a species. Several recent studies of the SARS-CoV-2 genome have identified signals of conservation and positive selection within the gene encoding Spike protein based on the ratio of synonymous to nonsynonymous substitution. Such tests cannot, however, detect changes in the function of RNA molecules.
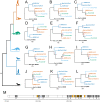
Methods: Here we apply a test for branch-specific oversubstitution of mutations within narrow windows of the genome without reference to the genetic code.
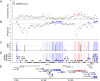
Results: We recapitulate the finding that the gene encoding Spike protein has been a target of both purifying and positive selection. In addition, we find other likely targets of positive selection within the genome of SARS-CoV-2, specifically within the genes encoding Nsp4 and Nsp16. Homology-directed modeling indicates no change in either Nsp4 or Nsp16 protein structure relative to the most recent common ancestor. These SARS-CoV-2-specific mutations may affect molecular processes mediated by the positive or negative RNA molecules, including transcription, translation, RNA stability, and evasion of the host innate immune system. Our results highlight the importance of considering mutations in viral genomes not only from the perspective of their impact on protein structure, but also how they may impact other molecular processes critical to the viral life cycle.
Keywords: COVID-19; Evolution; Genome; Positive selection; SARS-CoV-2.
© 2020 Berrio et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

