Pharmacological Inhibition of PPARy Boosts HIV Reactivation and Th17 Effector Functions, While Preventing Progeny Virion Release and de novo Infection
- PMID: 33089034
- PMCID: PMC7556414
- DOI: 10.20411/pai.v5i1.348
Pharmacological Inhibition of PPARy Boosts HIV Reactivation and Th17 Effector Functions, While Preventing Progeny Virion Release and de novo Infection
Abstract
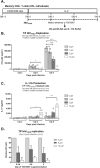
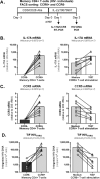
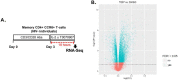
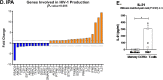
The frequency and functions of Th17-polarized CCR6+RORyt+CD4+ T cells are rapidly compromised upon HIV infection and are not restored with long-term viral suppressive antiretroviral therapy (ART). In line with this, Th17 cells represent selective HIV-1 infection targets mainly at mucosal sites, with long-lived Th17 subsets carrying replication-competent HIV-DNA during ART. Therefore, novel Th17-specific therapeutic interventions are needed as a supplement of ART to reach the goal of HIV remission/cure. Th17 cells express high levels of peroxisome proliferator-activated receptor gamma (PPARy), which acts as a transcriptional repressor of the HIV provirus and the rorc gene, which encodes for the Th17-specific master regulator RORyt. Thus, we hypothesized that the pharmacological inhibition of PPARy will facilitate HIV reservoir reactivation while enhancing Th17 effector functions. Consistent with this prediction, the PPARy antagonist T0070907 significantly increased HIV transcription (cell-associated HIV-RNA) and RORyt-mediated Th17 effector functions (IL-17A). Unexpectedly, the PPARy antagonism limited HIV outgrowth from cells of ART-treated people living with HIV (PLWH), as well as HIV replication in vitro. Mechanistically, PPARy inhibition in CCR6+CD4+ T cells induced the upregulation of transcripts linked to Th17-polarisation (RORyt, STAT3, BCL6 IL-17A/F, IL-21) and HIV transcription (NCOA1-3, CDK9, HTATIP2). Interestingly, several transcripts involved in HIV-restriction were upregulated (Caveolin-1, TRIM22, TRIM5α, BST2, miR-29), whereas HIV permissiveness transcripts were downregulated (CCR5, furin), consistent with the decrease in HIV outgrowth/replication. Finally, PPARy inhibition increased intracellular HIV-p24 expression and prevented BST-2 downregulation on infected T cells, suggesting that progeny virion release is restricted by BST-2-dependent mechanisms. These results provide a strong rationale for considering PPARy antagonism as a novel strategy for HIV-reservoir purging and restoring Th17-mediated mucosal immunity in ART-treated PLWH.
Keywords: ART; CD4+ T cells; HIV-1; IL-21; PPARy; Th17.
© Pathogens and Immunity 2020.
Conflict of interest statement
DP, AF, YZ, JPG, JR, AF, MJR, LRM, DC, HC, TRWS, and AG declare no financial or non-financial competing interests to disclose. EAC is a member of the Scientific Advisory Board of Theratechnologies. JPR performed contract research and/or served on Advisory Boards for Gilead Sciences Canada Inc., Merck Canada Inc., Abbvie Corp., ViiV Healthcare, Bristol Myers Squibb, Janssen Inc., Argos Pharmaceuticals from InnaVirVax, and Theravectys. NC received research funding from EMD Serono and served on Advisory Boards for Gilead Sciences. PA's laboratory receives research funding from Glaxo Smith Klein/NeoMed for projects different from the present study. PA served as a Consultant at Merck Canada Inc.
Figures











Similar articles
-
IL-17A reprograms intestinal epithelial cells to facilitate HIV-1 replication and outgrowth in CD4+ T cells.iScience. 2021 Oct 7;24(11):103225. doi: 10.1016/j.isci.2021.103225. eCollection 2021 Nov 19. iScience. 2021. PMID: 34712922 Free PMC article.
-
Impaired Th17 polarization of phenotypically naive CD4(+) T-cells during chronic HIV-1 infection and potential restoration with early ART.Retrovirology. 2015 Apr 30;12:38. doi: 10.1186/s12977-015-0164-6. Retrovirology. 2015. PMID: 25924895 Free PMC article.
-
Altered memory CCR6+ Th17-polarised T-cell function and biology in people with HIV under successful antiretroviral therapy and HIV elite controllers.EBioMedicine. 2024 Sep;107:105274. doi: 10.1016/j.ebiom.2024.105274. Epub 2024 Aug 22. EBioMedicine. 2024. PMID: 39178742 Free PMC article.
-
Th17 CD4+ T-Cell as a Preferential Target for HIV Reservoirs.Front Immunol. 2022 Feb 7;13:822576. doi: 10.3389/fimmu.2022.822576. eCollection 2022. Front Immunol. 2022. PMID: 35197986 Free PMC article. Review.
-
New Th17-specific therapeutic strategies for HIV remission.Curr Opin HIV AIDS. 2019 Mar;14(2):85-92. doi: 10.1097/COH.0000000000000522. Curr Opin HIV AIDS. 2019. PMID: 30543544 Review.
Cited by
-
A Canadian Survey of Research on HIV-1 Latency-Where Are We Now and Where Are We Heading?Viruses. 2024 Feb 1;16(2):229. doi: 10.3390/v16020229. Viruses. 2024. PMID: 38400005 Free PMC article. Review.
-
Retinoic acid enhances HIV-1 reverse transcription and transcription in macrophages via mTOR-modulated mechanisms.Cell Rep. 2024 Jul 23;43(7):114414. doi: 10.1016/j.celrep.2024.114414. Epub 2024 Jun 28. Cell Rep. 2024. PMID: 38943643 Free PMC article.
-
Metformin Enhances Antibody-Mediated Recognition of HIV-Infected CD4+ T-Cells by Decreasing Viral Release.bioRxiv [Preprint]. 2024 Feb 19:2024.02.15.580166. doi: 10.1101/2024.02.15.580166. bioRxiv. 2024. PMID: 38464135 Free PMC article. Preprint.
-
Genes involved in immune, gene translation and chromatin organization pathways associated with Mycoplasma ovipneumoniae presence in nasal secretions of domestic sheep.PLoS One. 2021 Jul 12;16(7):e0247209. doi: 10.1371/journal.pone.0247209. eCollection 2021. PLoS One. 2021. PMID: 34252097 Free PMC article.
-
Identification of aryl hydrocarbon receptor as a barrier to HIV-1 infection and outgrowth in CD4+ T cells.Cell Rep. 2023 Jun 27;42(6):112634. doi: 10.1016/j.celrep.2023.112634. Epub 2023 Jun 12. Cell Rep. 2023. PMID: 37310858 Free PMC article.
References
-
- Deeks SG, Lewin SR, Ross AL, Ananworanich J, Benkirane M, Cannon P, Chomont N, Douek D, Lifson JD, Lo YR, Kuritzkes D, Margolis D, Mellors J, Persaud D, Tucker JD, Barre-Sinoussi F, International ASTaCWG, Alter G, Auerbach J, Autran B, Barouch DH, Behrens G, Cavazzana M, Chen Z, Cohen EA, Corbelli GM, Eholie S, Eyal N, Fidler S, Garcia L, Grossman C, Henderson G, Henrich TJ, Jefferys R, Kiem HP, Mc-Cune J, Moodley K, Newman PA, Nijhuis M, Nsubuga MS, Ott M, Palmer S, Richman D, Saez-Cirion A, Sharp M, Siliciano J, Silvestri G, Singh J, Spire B, Taylor J, Tolstrup M, Valente S, van Lunzen J, Walensky R, Wilson I, Zack J.. International AIDS Society global scientific strategy: towards an HIV cure 2016. Nature medicine. 2016;22(8):839-50. Epub 2016/07/12. doi: 10.1038/nm.4108. PubMed PMID: 27400264; PMCID: PMC5322797. - DOI - PMC - PubMed
-
- Sengupta S, Siliciano RF.. Targeting the Latent Reservoir for HIV-1. Immunity. 2018;48(5):872-95. Epub 2018/05/17. doi: 10.1016/j.immuni.2018.04.030. PubMed PMID: 29768175; PMCID: PMC6196732. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
