Erythropoietin Regulates Transcription and YY1 Dynamics in a Pre-established Chromatin Architecture
- PMID: 33089097
- PMCID: PMC7559257
- DOI: 10.1016/j.isci.2020.101583
Erythropoietin Regulates Transcription and YY1 Dynamics in a Pre-established Chromatin Architecture
Abstract
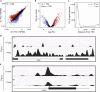
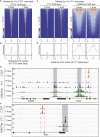
The three-dimensional architecture of the genome plays an essential role in establishing and maintaining cell identity. However, the magnitude and temporal kinetics of changes in chromatin structure that arise during cell differentiation remain poorly understood. Here, we leverage a murine model of erythropoiesis to study the relationship between chromatin conformation, the epigenome, and transcription in erythroid cells. We discover that acute transcriptional responses induced by erythropoietin (EPO), the hormone necessary for erythroid differentiation, occur within an invariant chromatin topology. Within this pre-established landscape, Yin Yang 1 (YY1) occupancy dynamically redistributes to sites in proximity of EPO-regulated genes. Using HiChIP, we identify chromatin contacts mediated by H3K27ac and YY1 that are enriched for enhancer-promoter interactions of EPO-responsive genes. Taken together, these data are consistent with an emerging model that rapid, signal-dependent transcription occurs in the context of a pre-established chromatin architecture.
Keywords: Developmental Biology; Genomics; Molecular Biology.
© 2020 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Epo reprograms the epigenome of erythroid cells.Exp Hematol. 2017 Jul;51:47-62. doi: 10.1016/j.exphem.2017.03.004. Epub 2017 Apr 12. Exp Hematol. 2017. PMID: 28410882 Free PMC article.
-
YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment.Genome Res. 2017 Jul;27(7):1139-1152. doi: 10.1101/gr.215160.116. Epub 2017 May 23. Genome Res. 2017. PMID: 28536180 Free PMC article.
-
Integrative view on how erythropoietin signaling controls transcription patterns in erythroid cells.Curr Opin Hematol. 2018 May;25(3):189-195. doi: 10.1097/MOH.0000000000000415. Curr Opin Hematol. 2018. PMID: 29389768 Free PMC article. Review.
-
The Why of YY1: Mechanisms of Transcriptional Regulation by Yin Yang 1.Front Cell Dev Biol. 2020 Sep 30;8:592164. doi: 10.3389/fcell.2020.592164. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33102493 Free PMC article. Review.
-
In Vivo Chromatin Targets of the Transcription Factor Yin Yang 2 in Trophoblast Stem Cells.PLoS One. 2016 May 18;11(5):e0154268. doi: 10.1371/journal.pone.0154268. eCollection 2016. PLoS One. 2016. PMID: 27191592 Free PMC article.
Cited by
-
YY1-controlled regulatory connectivity and transcription are influenced by the cell cycle.Nat Genet. 2024 Sep;56(9):1938-1952. doi: 10.1038/s41588-024-01871-y. Epub 2024 Aug 29. Nat Genet. 2024. PMID: 39210046 Free PMC article.
-
YY1 knockout in pro-B cells impairs lineage commitment, enabling unusual hematopoietic lineage plasticity.Genes Dev. 2024 Oct 16;38(17-20):887-914. doi: 10.1101/gad.351734.124. Genes Dev. 2024. PMID: 39362773 Free PMC article.
-
STAT5 as a Key Protein of Erythropoietin Signalization.Int J Mol Sci. 2021 Jul 1;22(13):7109. doi: 10.3390/ijms22137109. Int J Mol Sci. 2021. PMID: 34281163 Free PMC article. Review.
References
-
- Atlasi Y., Megchelenbrink W., Peng T., Habibi E., Joshi O., Wang S.Y., Wang C., Logie C., Poser I., Marks H., Stunnenberg H.G. Epigenetic modulation of a hardwired 3D chromatin landscape in two naive states of pluripotency. Nat. Cell Biol. 2019;21:568–578. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

